Abstract
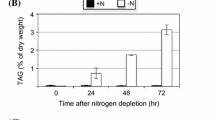
Autophagy mediates degradation and recycling of cellular components and plays an important role in senescence and adaptive responses to biotic and abiotic stresses. Nutrient deprivation has been shown to trigger triacylglycerol (TAG) accumulation and also induces autophagy in various green algae. However, the functional relationship between TAG metabolism and autophagy remains unclear. To gain preliminary evidence supporting a role of autophagy in TAG synthesis, Chlamydomonas reinhardtii CC-2686 was grown in Tris-acetate phosphate medium with or without nitrogen and treated with an autophagy inducer (rapamycin) or inhibitors (wortmannin, 3-methyladenine, and bafilomycin A1). Fluorescence microscopic analysis of Nile red-stained cells following 72-h treatments showed that rapamycin induced accumulation of subcellular lipid droplets which are storage sites of TAG. Rapamycin treatment in combination with nitrogen starvation led to a greater abundance of lipid droplets. Wortmannin and bafilomycin A1, but not 3-methyladenine, inhibited lipid droplet accumulation in rapamycin-treated cells and to a less extent in nitrogen-depleted cells. These results suggested that autophagy contributes to TAG synthesis in C. reinhardtii, but is not a necessary process. Autophagy induction may also be used to further enhance TAG accumulation in microalgae under nutrient deprivation.




Similar content being viewed by others
References
Ball S, Marianne T, Dirick L, Fresnoy M, Delrue B, Decq A (1991) A Chlamydomonas reinhardtii low-starch mutant is defective for 3-phosphoglycerate activation and orthophosphate inhibition of ADP-glucose pyrophosphorylase. Planta 185:17–26
Bassham DC (2007) Plant autophagy-more than a starvation response. Curr Opin Plant Biol 10:587–593
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287:15811–15825
Brauer D, Uknalis J, Triana R, Shachar-Hill Y, Tu S-I (1997) Effects of bafilomycin A1 and metabolic inhibitors on the maintenance of vacuolar acidity in maize root hair cells. Plant Physiol 113:809–816
Crespo JL, Díaz-Troya S, Florencio FJ (2005) Inhibition of target of rapamycin signaling by rapamycin in the unicellular green alga Chlamydomonas reinhardtii. Plant Physiol 139:1736–1749
Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL (2008) The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4:851–865
Ehara M, Noguchi T, Ueda K (1996) Uptake of neutral red by the vacuoles of a green alga, Micrasterias pinnatifida. Plant Cell Physiol 37:734–741
Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585:1985–1991
Fingar DC, Blenis J (2004) Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151–3171
Goold H, Beisson F, Peltier G, Li-Beisson Y (2015) Microalgal lipid droplets: composition, diversity, biogenesis and functions. Plant Cell Rep 34:545–555
Imamura S, Kawase Y, Kobayashi I, Sone T, Era A, Miyagishima SY, Shimojima M, Ohta H, Tanaka K (2015) Target of rapamycin (TOR) plays a critical role in triacylglycerol accumulation in microalgae. Plant Mol Biol 89:309–318
Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47:1641–1652
Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C (2002) Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc Natl Acad Sci U S A 99:6422–6427
Merchant SS, Kropat J, Liu B, Shaw J, Warakanont J (2012) TAG, You’re it! Chlamydomonas as a reference organism for understanding algal triacylglycerol accumulation. Curr Opin Biotech 23:352–363
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140:313–326
Pérez-Pérez ME, Florencio FJ, Crespo JL (2010) Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol 152:1874–1888
Pérez-Pérez ME, Lemaire SD, Crespo JL (2012) Reactive oxygen species and autophagy in plants and algae. Plant Physiol 160:156–164
Ren C, Liu J, Gong Q (2014) Functions of autophagy in plant carbon and nitrogen metabolism. Front Plant Sci 5:301. doi:10.3389/fpls.2014.00301
Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ (2007) Potential therapeutic applications of autophagy. Nat Rev Drug Discov 6:304–312
Scott RC, Juhász G, Neufeld TP (2007) Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17:1–11
Sirikhachornkit A, Vuttipongchaikij S, Suttangkakul A, Yokthongwattana K, Juntawong P, Pokethitiyook P, Kangvansaichol K, Meetam M (2016) Increasing the triacylglycerol content in Dunaliella tertiolecta through isolation of starch-deficient mutants. J Microbiol Biotechnol 28:854–866
Takatsuka C, Inoue Y, Matsuoka K, Moriyasu Y (2004) 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 45:265–274
Wang Z, Benning C (2011) Arabidopsis thaliana polar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J Vis Exp 49:2518
Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U (2009) Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell 8:1856–1868
Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23:33–42
Zhang H, Zeng R, Chen D, Liu J (2016) A pivotal role of vacuolar H+-ATPase in regulation of lipid production in Phaeodactylum tricornutum. Sci Rep 6:31319. doi:10.1038/srep31319
Acknowledgements
This research was supported by the PTT Research and Technology Institute, the Royal Golden Jubilee Ph.D. Program of Thailand (Thailand Research Fund), the Center of Excellence on Environmental Health and Toxicology (EHT), the Science & Technology Postgraduate Education and Research Development Office (PERDO), and the Ministry of Education, Thailand.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Fig. S1
Appearance of C. reinhardtii CC-2686 cultures grown for 72 h in normal TAP medium (+N), TAP-N (-N), and TAP medium containing 1 μM rapamycin (+N, Rapa(1 μM)) in which autophagy inhibitors wortmannin (WM), 3-methyladenine (3-MA), and bafilomycin A1 (Baf) were added to the final concentration as indicated. (JPEG 169 kb)
Fig. S2
Representative images of Nile red-stained cells of C. reinhardtii CC-2686. The alga was grown in TAP-N medium, TAP-N medium containing inhibitors, and TAP-N medium containing 1 μM rapamycin for 72 h. Scale bar represents 10 μm. Three images are shown for each treatment. (JPEG 139 kb)
Fig. S3
TLC analysis of total lipids from C. reinhardtii CC-124 cells grown in normal TAP medium (+N), TAP medium containing 1 μM rapamycin (+N, Rapa(1 μM)), TAP medium containing 1 μM rapamycin together with 0.1 μM bafilomycin A1 (+N, Rapa(1 μM)-Baf(0.1 μM)), TAP-N medium (-N), and TAP-N medium containing 0.1 μM bafilomycin A1 (-N, Baf(0.1 μM) for 72 h. Equal amounts of cells, 5×106 cells, were used for each lane. Representative images are shown. (JPEG 56 kb)
Fig. S4
Fatty acid composition of TAG derived from TLC separation (Fig. S3) of total lipids from 5×106 cells C. reinhardtii CC-124 cells grown in normal TAP medium (+N), TAP medium containing 1 μM rapamycin (+N, Rapa(1 μM)), TAP medium containing 1 μM rapamycin together with 0.1 μM bafilomycin A1 (+N, Rapa(1 μM)-Baf(0.1 μM)), TAP-N medium (-N), and TAP-N medium containing 0.1 μM bafilomycin A1 (-N, Baf(0.1 μM) for 72 h. Bands corresponding to TAG were removed from TLC plates, transesterified with methanol, and quantified using GC-MS as described in materials and methods. Each bar represents mean ± SEM of 6 replications. (JPEG 52 kb)
Table S1
(DOCX 12 kb)
Rights and permissions
About this article
Cite this article
Pugkaew, W., Meetam, M., Ponpuak, M. et al. Role of autophagy in triacylglycerol biosynthesis in Chlamydomonas reinhardtii revealed by chemical inducer and inhibitors. J Appl Phycol 30, 15–22 (2018). https://doi.org/10.1007/s10811-017-1166-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-017-1166-7