Abstract
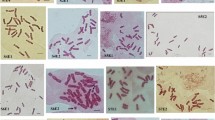
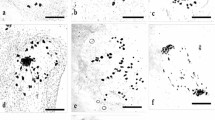
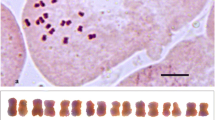
Diploids, triploids and tetraploids were screened from the germplasm garden of Chinese chive in this study using flow cytometry and chromosome counting technologies. Their evolutionary status was analyzed by karyotype comparison and microsporogenesis observation. Their relationships were determined and discussed. The karyotype formula of the diploids was 2n = 2x = 16 = 14 m + 2sm (2SAT). The asymmetrical karyotype coefficient (As. K. C) was 57.02%. The karyotype formula of triploids was 2n = 3x = 24 = 18 m + 6sm (2SAT), and the As. K. C was 57.49%. The karyotype formula of tetraploids was 2n = 4x = 32 = 26 m + 4sm + 2st (2SAT), and the As. K. C was 57.80%. The karyoty** results suggested that the evolutionary status increased with increasing ploidy in Chinese chive, although all of them were 2A karyotypes. Abnormal chromosome synapsis and separation during microsporogenesis result in the formation of pollen of different sizes in triploids, suggesting that 2n gametes more easily form in triploids. Therefore, we speculate that tetraploids were formed by the combination of 2n gametes of a triploid and n gametes of a diploid. In this process, triploids are the most likely bridge for the formation of tetraploids in Chinese chive.






Similar content being viewed by others
Data availability
All the data are available in the submitted manuscript.
References
Arano H (1963) Cytological studies in subfamily carduoideae (Compositae) of Japan. IX Bot Mag (tokyo) 76:32–39
Bretagnolle F, Thompson JD (1995) Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol 129:1–22. https://doi.org/10.1111/j.1469-8137.1995.tb03005.x
Cao Y, Zhao K, Xu J et al (2023) Genome balance and dosage effect drive allopolyploid formation in Brassica. PNAS 120(14):e2217672120. https://doi.org/10.1073/pnas.2217672120
Edet OU, Aikpokpodion PO (2014) Karyotype analysis of Ocimum basilicum in South-Eastern Nigeria. Am J Plant Sci 5:126–131. https://doi.org/10.4236/ajps.2014.51016
Fox DT, Soltis DE, Soltis PS, Ashman TL, Van de Peer Y (2020) Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol 30(9):688–694. https://doi.org/10.1016/j.tcb.2020.06.006
Harlan JR, De Wet JMJ (1975) On Ö Winge and a prayer: the origins of polyploidy. Bot Rev 41:361–390. https://doi.org/10.1007/BF02860830
Huang YX, Yu PC, Zhang Q, Yue ZY, Piao ML, Gao X, Jia GX (2023) Effective induction of fertile gametes in oriental × trumpet (OT) lily by high temperatures. Plants 12(7):1563. https://doi.org/10.3390/plants12071563
Jiang K, Kuang Y, Feng L, Liu Y, Wang S, Du H, Shen W (2021) Molecular hydrogen maintains the storage quality of Chinese chive through improving antioxidant capacity. Plants 10(6):1095. https://doi.org/10.3390/plants10061095
Jiao ZY, Yin SH, Chen JH (2022) Analysis on floral phenotypic diversity of Chinese chive germplasm resources China. Vegetables 404(10):66–73
Kojima A, Nagato Y (1997) Discovery of highly apomictic and highly amphimictic dihaploids in Allium tuberosum. Sex Plant Reprod 10:8–12. https://doi.org/10.1007/s004970050061
Lavia GI, Ortiz AM, Robledo G, Fernández A, Seijo G (2011) Origin of triploid Arachis pintoi (Leguminosae) by autopolyploidy evidenced by FISH and meiotic behaviour. Ann Bot 108(1):103–111. https://doi.org/10.1093/aob/mcr108
Li MX, Shang ST (1982) An investigation on the karyotype of Allium tuberosum Rottler and A. ramosum L. Acta Horticulturae Sinica 9(3):31–36
Li Y, Zhang H, Cui Y, Chen J, Lü A, Li J, Li Y (2020) Analysis on SSR information in full-length transcriptome and development of molecular markers in Allium tuberosum. Acta Horticulturae Sinica 47(04):759–768. https://doi.org/10.16420/j.issn.0513-353x.2019-0589
Liu N, Tong J, Hu M, Ji Y, Wang B, Liang H, Liu M, Wu Z (2021) Transcriptome landscapes of multiple tissues highlight the genes involved in the flavor metabolic pathway in Chinese chive (Allium tuberosum). Genomics 113:2145–2157. https://doi.org/10.1016/j.ygeno.2021.05.005
Mahoney JD, Hau TM, Connolly BA, Brand MH (2019) Sexual and apomictic seed reproduction in Aronia species with different ploidy levels. HortScience 54(4):642–646. https://doi.org/10.21273/HORTSCI13772-18
Mathur M, Tandon SL (1965) Cytology of an autotetraploid Allium tuberosum. Ind J Hort 22:382–384
Pedro MR, Delgado-Salinas A (2009) Karyotypic analysis in six species of Phaseolus L. (Fabaceae). Caryologia 62(3):167–170
Peruzzi L, Eroğlu HE (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet 7(1):1. https://doi.org/10.3897/CompCytogen.v7i1.4431
Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol S 29:467–501. https://doi.org/10.1146/annurev.ecolsys.29.1.467
Rieseberg LH, Willis JH (2007) Plant speciation. Science 317(5840):910–914. https://doi.org/10.1126/science.1137729
Sartor ME, Quarin CL, Urbani MH, Espinoza F (2011) Ploidy levels and reproductive behaviour in natural populations of five paspalum species. Plant Syst and Evol 293:31–41. https://doi.org/10.1007/s00606-011-0416-4
Session AM, Uno Y, Kwon T et al (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538(7625):336–343. https://doi.org/10.1038/nature19840
Sharma G, Gohil RN (2013) Origin and cytology of a novel cytotype of Allium tuberosum Rottl. ex Spreng. (2n = 48). Genet Resour Crop Evol 60:503–511. https://doi.org/10.1007/s10722-012-9852-4
Stebbins GL (1971) Chromosomal evolution in higher plants. Chromosomal evolution in higher plants
Tong J, Hu M, Han B, Ji Y, Wang B, Liang H, Liu M, Wu Z, Liu N (2021) Determination of reliable reference genes for gene expression studies in Chinese chive (Allium tuberosum) based on the transcriptome profiling. Sci Rep 11:16558. https://doi.org/10.1038/s41598-021-95849-z
Van de Peer Y, Ashman TL, Soltis PS, Soltis DE (2021) Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33(1):11–26. https://doi.org/10.1093/plcell/koaa015
Van Tuyl JM, Lim KB (2003) Interspecific hybridisation and polyploidisation as tools in ornamental plant breeding. Acta Hort 612:13–22. https://doi.org/10.17660/ActaHortic.2003.612.1
**e B, Wu Q, Wei S, Li H, Wei J, Hanif M, Li J, Liu Z, **ao X, Yu J (2022) Optimization of headspace solid-phase micro-extraction conditions (HS-SPME) and identification of major volatile aroma-active compounds in Chinese chive (Allium tuberosum Rottler). Molecules 27:2425. https://doi.org/10.3390/molecules27082425
Yamashita K, Nakazawa Y, Namai K, Amagai M, Tsukazaki H, Wako T, Kojima A (2012) Modes of inheritance of two apomixis components, diplospory and parthenogenesis, in Chinese chive (Allium ramosum) revealed by analysis of the segregating population generated by back-crossing between amphimictic and apomictic diploids. Breed Sci 62:160–169. https://doi.org/10.1270/jsbbs.62.160
Yan Y (1986) The karyotype, meiotic characteristics and potential value in cultivation of a triploid of Allium tuberosum. J Yunnan Univ 8(4):422–427
Yang Y, Chen Z (1994) Karyotype analysis of wild Allium tuberosum. J Guangdong Med Coll 02:143–144
Yang L, Xu JM, Zhang XL, Wan HQ (1998) Karyotypical studies of six species on the genus Allium. Acta Phytotaxonomica Sinica 36(1):36–46
Yao PQ, Li GH, Qiu YF, Kang XY (2020) Induction of 2n female gametes in rubber (Hevea brasiliensis) by high-temperature exposure during megasporogenesis as a basis for triploid breeding. Tree Genet Genomes 16(1):24. https://doi.org/10.1007/s11295-020-1413-y
Zhou Q, Cheng X, Kong B, Zhao Y, Li Z, Sang Y, Wu J, Zhang P (2022) Heat shock-induced failure of meiosis I to meiosis II transition leads to 2n pollen formation in a woody plant. Plant Physiol 189(4):2110–2127. https://doi.org/10.1093/plphys/kiac219
Funding
This work was supported by the Scientific and Technology Project in Henan Province, China (212102110040) and Graduate Program Funding of Henan University of Science and Technology (2020YJG-020).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Peng-Qiang Yao, Jian-Hua Chen, Pei-Fang Ma and Li-Hua **e. The first draft of the manuscript was written by Peng-Qiang Yao. Jiang Shui and Shi-** Cheng commented on previous versions of the manuscript. All the authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yao, PQ., Chen, JH., Ma, PF. et al. Karyotype analysis of Chinese chive germplasms with different ploidy levels and their evolutionary relationships. Genet Resour Crop Evol 71, 1749–1758 (2024). https://doi.org/10.1007/s10722-023-01717-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01717-2