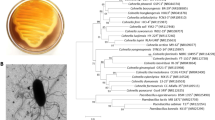
Abstract
Objective
To identify and characterize a new β-agarase from Cellulophaga omnivescoria W5C capable of producing biologically-active neoagarooligosaccharides from agar.
Results
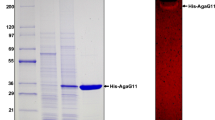
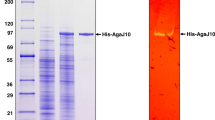
The β-agarase, Aga1, has signal peptides on both N- and C-terminals, which are involved in the type IX secretion system. It shares 75% protein sequence identity with AgaD from Zobellia galactanivorans and has a molecular weight of 54 kDa. Biochemical characterization reveals optimum agarolytic activities at pH 7–8 and temperature 30–45 °C. Aga1 retains at least 33% activity at temperatures lower than the sol–gel transition state of agarose. Metal ions are generally not essential, but calcium and potassium enhance its activity whereas iron and zinc are inhibitory. Finally, hydrolysis of agarose with Aga1 yields neoagarotetraose, neoagarohexaose, and neoagarooctaose.
Conclusions
Aga1 displays unique traits such as moderate psychrophilicity, stability, and synergy with other agarases, which makes it an excellent candidate for biosynthetic production of neoagarooligosaccharides from agar.




Similar content being viewed by others
References
An K, Shi X, Cui F, Cheng J, Liu N, Zhao X, Zhang X-H (2018) Characterization and overexpression of a glycosyl hydrolase family 16 beta-agarase YM01-1 from marine bacterium Catenovulum agarivorans YM01T. Protein Expr Purif 143:1–8. https://doi.org/10.1016/j.pep.2017.10.002
Chi W-J, Lee C, Dugerjonjuu S, Park J-S, Kang D-K, Hong S-K (2015) Biochemical characterization of a novel iron-dependent GH16 β-agarase, AgaH92, from an agarolytic bacterium Pseudoalteromonas sp. H9. FEMS Microbiol Lett 362:fnv35. https://doi.org/10.1093/femsle/fnv035
Chi W, Chang Y, Hong S (2012) Agar degradation by microorganisms and agar-degrading enzymes. Appl Microbiol Biotechnol 94:917–930. https://doi.org/10.1007/s00253-012-4023-2
Fu X, Pan C-H, Lin H, Kim S (2009) Gene cloning, expression, and characterization of a β-agarase, agaB34, from Agarivorans albus YKW-34. J Microbiol Biotechnol 19:257–264
Fu XT, Kim SM (2010) Agarase: review of major sources, categories, purification method, enzyme characteristics and applications. Marine Drugs. https://doi.org/10.3390/md8010200
Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C, Porro D, Ferrer P, Tutino ML, Mattanovich D, Villaverde A (2008) Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact 7:11. https://doi.org/10.1186/1475-2859-7-11
Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A (2005) Protein Identification and Analysis Tools on the ExPASy Server. In: Walker JM (ed) The Proteomics Protocols Handbook. Humana Press, New Jersey, pp 571–607. https://doi.org/10.1385/1-59259-890-0:571
Hehemann J-H, Correc G, Thomas F, Bernard T, Barbeyron T, Jam M, Helbert W, Michel G, Czjzek M (2012a) Biochemical and structural characterization of the complex agarolytic enzyme system from the marine bacterium Zobellia galactanivorans. J Biol Chem 287:30571–30584. https://doi.org/10.1074/jbc.m112.377184
Hehemann J-H, Kelly AG, Pudlo NA, Martens EC, Boraston AB (2012b) Bacteria of the human gut microbiome catabolize red seaweed glycans with carbohydrate-active enzyme updates from extrinsic microbes. Proc National Acad Sci USA 109:19786. https://doi.org/10.1073/pnas.1211002109
Jam M, Flament D, Allouch J, Potin P, Thion L, Kloareg B, Czjzek M, Helbert W, Michel G, Barbeyron T (2005) The endo-β-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: two paralogue enzymes with different molecular organizations and catalytic behaviours. Biochem J 385:703–713. https://doi.org/10.1042/bj20041044
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Kobayashi R, Takisada M, Suzuki T, Kirimura K, Usami S (1997) Neoagarobiose as a novel moisturizer with whitening effect. Biosci Biotechnol Biochem 61:162–163
Lasica AM, Goulas T, Mizgalska D, Zhou X, de Diego I, Ksiazek M, Madej M, Guo Y, Guevara T, Nowak M, Potempa B, Goel A, Sztukowska M, Prabhakar AT, Bzowska M, Widziolek M, Thøgersen IB, Enghild JJ, Simonian M, Kulczyk AW, Nguyen K-A, Potempa J, Gomis-Rüth FX (2016) Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. Sci Rep 6:37708. https://doi.org/10.1038/srep37708
Malleshappa Gowder S, Chatterjee J, Chaudhuri T, Paul K (2014) Prediction and analysis of surface hydrophobic residues in tertiary structure of proteins. Sci World J 2014:971258. https://doi.org/10.1155/2014/971258
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Petersen T, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. https://doi.org/10.1038/nmeth.1701
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Ramos KRM, Valdehuesa KNG, Cabulong RB, Moron LS, Nisola GM, Hong S-K, Lee W-K, Chung W-J (2016) Overexpression and secretion of AgaA7 from Pseudomonas hodoensis sp. nov in Bacillus subtilis for depolymerization of agarose. Enzyme Microb Technol 90:19–25
Ramos KRM, Valdehuesa KNG, Maza PAMM, Nisola GM, Lee W-K, Chung W-J (2017) Overexpression and characterization of a novel α-neoagarobiose hydrolase and its application in the production of D-galactonate from Gelidium amansii. Process Biochem 63:105–112
Ramos KRM, Valdehuesa KNG, Nisola GM, Lee W-K, Chung W-J (2018) Identification and characterization of a thermostable endolytic β-agarase Aga2 from a newly isolated marine agarolytic bacteria Cellulophaga omnivescoria W5C. New Biotechnol 40:261–267
Rochas C, Lahaye M, Yaphe W, Viet MTP (1986) 13C-NMR-spectroscopic investigation of agarose oligomers. Carbohydr Res. https://doi.org/10.1016/S0008-6215(00)90388-4
Strub C, Alies C, Lougarre A, Ladurantie C, Czaplicki J, Fournier D (2004) Mutation of exposed hydrophobic amino acids to arginine to increase protein stability. BMC Biochem 2(5):9. https://doi.org/10.1186/1471-2091-5-9
Temuu** U, Chi W-J, Lee S-Y, Chang Y-K, Hong S-K (2011) Overexpression and biochemical characterization of DagA from Streptomyces coelicolor A3(2): an endo-type β-agarase producing neoagarotetraose and neoagarohexaose. Appl Microbiol Biotechnol 92:749–759. https://doi.org/10.1007/s00253-011-3347-7
Valdehuesa KNG, Ramos KRM, Moron LS, Lee I, Nisola GM, Lee W-K, Chung W-J (2018) Draft genome sequence of newly isolated agarolytic bacteria Cellulophaga omnivescoria sp. nov. W5C carrying several gene loci for marine polysaccharide degradation. Curr Microbiol 75:925–933. https://doi.org/10.1007/s00284-018-1467-3
Vazquez E, Corchero JL, Villaverde A (2011) Post-production protein stability: trouble beyond the cell factory. Microb Cell Fact 10:60. https://doi.org/10.1186/1475-2859-10-60
Veerakumar S, Manian RP (2018) Recombinant β-agarases: insights into molecular, biochemical, and physiochemical characteristics. 3 Biotech 8(10):445. https://doi.org/10.1007/s13205-018-1470-1
Vogt G, Woell S, Argos P (1997) Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 269:631–643. https://doi.org/10.1006/jmbi.1997.1042
Wass MN, Kelley LA, Sternberg MJ (2010) 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res 38:W469–W473. https://doi.org/10.1093/nar/gkq406
Supporting information
Supplementary Table 1—Primers used for the PCR amplification of full and truncated aga1 gene from Cellulophaga omnivescoria sp. nov. W5C.
Supplementary Fig. 1—Amino acid sequence alignment of Aga1 (Aga1-Co) with some known GH16 β-agarases. AgaD (Acc. No. CAZ98378.1), AgaB (Acc. No. CAZ97711.1), and AgaA (Acc. No. CAZ98338.1) from Zobellia galactanivorans; BpGH16A (Acc. No. EDY95404.1); AgaB34 (Acc. No. ABW77762.1) from Agarivorans albus; and DagA (Acc. No. NP_627674.1) from Streptomyces coelicolor. Black and white arrowheads denote N- and C-terminal signal peptide cleavage sites, respectively. Green and purple coils represent α- and transmembrane helices, respectively. Blue arrows indicate β-sheets. Red squares denote catalytic residues. Closed and open circles represent conserved and predicted substrate binding residues, respectively. Multiple sequence alignment and visualization were carried out using CLC Sequence Viewer 8.0.
Supplementary Fig. 2—Supernatant activity assay. The enzyme assay was performed using the optimum conditions. FL for expression of full length Aga1 and (-)N,C for N- and C- terminal truncated Aga1.
Supplementary Fig. 3—Effect of repeated freezing-thawing on Aga1 activity. The enzyme assay was performed using the optimum conditions.
Supplementary Fig. 4—(A) Molecular plot of the hydrophobic surface of Aga1 (The color of the surface indicates the hydrophobicity ranges from dark goldenrod (most hydrophobic), then white (moderate), to dark cyan (most hydrophilic). (B) Predicted solvent accessibility of the amino acids in Aga1 (Values range from 0 for buried residue to 9 for highly exposed residue). The data showed here were acquired using I-TASSER (Iterative Threading Assembly Refinement) online tool and the protein structure was polished using ChimeraX software (Yang and Zhang, 2015; Zhang et al. 2017; Goddard et al. 2018).
Supplementary Fig. 5—13C-NMR spectra of hydrolysis products.
Supplementary Fig. 6—3D structural modelling of Aga1. (A) Superimposed Aga1 (blue) and AgaD (red). (B) Hydrophobic (orange), negatively-charged (blue), and positively-charged (green) residues.
Funding
This work was supported by Korea Research Fellowship Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (Grant No. 2015H1D3A1062172) and by the Ministry of Education (Grant No. 2018R1D1A1B07043993) and by the Korea Institute of Technology Evaluation and Planning (KETEP) funded by the Ministry of Trade, Industry & Energy (Grant No. MOTIE 20194010201750).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no competing interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ramos, K.R.M., Valdehuesa, K.N.G., Bañares, A.B. et al. Overexpression and characterization of a novel GH16 β-agarase (Aga1) from Cellulophaga omnivescoria W5C. Biotechnol Lett 42, 2231–2238 (2020). https://doi.org/10.1007/s10529-020-02933-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02933-x