Abstract
Objectives
2,3-Butanediol (2,3-BD) is widely used in several chemical syntheses as well as the manufacture of plastics, solvents, and antifreeze formulations, and can be manufactured by microbial glucose fermentation. Conventional (2,3-BD) fermentation typically has low productivity, yield, and purity, and is expensive for commercial applications. We aimed to delete the lactate dehydrogenase and acetate kinase (ldhA and ack) genes in Klebsiella pneumoniae HD79 by using λRed homologous recombination technology, to eliminate by-products and thereby improve (2,3-BD) production. We also analyzed the resulting gene changes by using transcriptomics.
Results
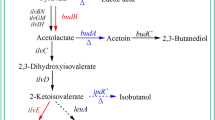
The yield of (2,3-BD) from the mutant Klebsiella strain was 46.21 g/L, the conversion rate was 0.47 g/g, and the productivity was 0.64 g/L·h, which represented increases of 54.9%, 20.5%, and 106.5% respectively, compared to (WT) strains. Lactate and acetate decreased by 48.2% and 62.8%, respectively. Transcriptomics analysis showed that 4628 genes were differentially expressed (404 significantly up-regulated and 162 significantly down-regulated). Moreover, the (2,3-BD) operon genes were differentially expressed.
Conclusion
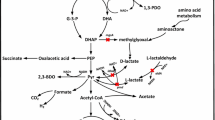
Our data showed that the biosynthesis of (2,3-BD) was regulated by inducers (lactate and acetate), a regulator (BudR), and carbon flux. Elimination of acidic by-products by ldhA and ack knockdown significantly improved (2,3-BD) production. Our results provide a deeper understanding of the mechanisms underlying (2,3-BD) production, and form a molecular basis for the improvement this process by genetic modification in the future.




Similar content being viewed by others
References
Białkowska AM (2016) Strategies for efficient and economical 2,3-butanediol production: new trends in this field. World J Microbiol Biotechnol 32:1–14
Chen L, Ma C, Wang R, Yang J, Zheng H (2016) Deletion of ldhA and aldH genes in Klebsiella pneumoniae to enhance 1,3-propanediol production. Biotechnol Lett 38:1769–1774
Dimont E, Shi J, Kirchner R, Hide W (2015) edgeRun: an R package for sensitive, functionally relevant differential expression discovery using an unconditional exact test. Bioinform 31:2589–2590
Förster AH, Gescher J (2014) Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Biotechnol Bioeng 2:16
Gao Y, Huang H, Chen S, Qi G (2018) Production of optically pure 2,3-butanediol from Miscanthus floridulus hydrolysate using engineered Bacillus licheniformis strains. World J Microbiol Biotechnol 34:66
Ge Y, Li K, Li L, Chao G, Zhang L, Ma C, ** X (2016) Contracted but effective: production of enantiopure 2,3-butanediol by thermophilic and GRAS Bacillus licheniformis. Green Chem 18:4693–4703
Guo X, Cao C, Wang Y, Li C, Wu M, Chen Y, Zhang C, Pei H, **ao D (2014) Effect of the inactivation of lactate dehydrogenase, ethanol dehydrogenase, and phosphotransacetylase on 2,3-butanediol production in Klebsiella pneumoniae strain. Biotechnol Biofuels 7:44
Guo XW, Zhang Y, Li LL, Guan XY, Guo J, Wu DG, Chen YF, **ao GD (2018) Improved xylose tolerance and 2,3-butanediol production of Klebsiella pneumoniae by directed evolution of rpoD and the mechanisms revealed by transcriptomics. Biotechnol Biofuels 11:307
Harney E, Dubief B, Boudry P, Basuyaux O, Schilhabel MB, Huchette S, Paillard C, Nunes FLD (2016) De novo assembly and annotation of the European abalone Haliotis tuberculata transcriptome. Mar Genom 28:11–16
Harvianto GR, Haider J, Hong J, Long NVD, Shim JJ, Cho MH, Kim WK, Lee M (2018) Purification of 2,3-butanediol from fermentation broth: process development and techno-economic analysis. Biotechnol Biofuels 11:18
Heum SS, Sewhan K, Jae Young K, Soo** L, Youngsoon U, Min-Kyu O, Young-Rok K, **won L (2012) Complete genome sequence of the 2,3-butanediol-producing Klebsiella pneumoniae strain KCTC 2242. J Bacteriol 194:2736–2737
Ishii J, Morita K, Ida K, Kato H, Kinoshita S, Hataya S, Shimizu H, Kondo A, Matsuda F (2018) A pyruvate carbon flux tugging strategy for increasing 2,3-butanediol production and reducing ethanol subgeneration in the yeast Saccharomyces cerevisiae. Biotechnol Biofuels 11:180
Ji XJ, Huang H, Li S, Du J, Lian M (2008) Enhanced 2,3-butanediol production by altering the mixed acid fermentation pathway in Klebsiella oxytoca. Biotechnol Lett 30:731–734
Ji XJ, Huang H, Ouyang PK (2011) Microbial 2,3-butanediol production: a state-of-the-art review. Biotechnol Adv 29:351–364
Kanehisa M, Furumichi M, Mao T, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361
Kim DK, Rathnasingh C, Song H, Lee HJ, Seung D, Yong KC (2013) Metabolic engineering of a novel Klebsiella oxytoca strain for enhanced 2,3-butanediol production. J Biosci Bioeng 116:186–192
Kiran Gopinath B, Vivek Nagaraj T, Rohit Nandan S, Madavan V (2015) Ameliorated de novo transcriptome assembly using illumina paired end sequence data with trinity assembler. Genomics 5:352–359
Lee S, Kim B, Jeong D, Oh M, Um Y, Kim YR, Kim J, Lee J (2013) Observation of 2,3-butanediol biosynthesis in Lys regulator mutated Klebsiella pneumoniae at gene transcription level. J Biotechnol 168:520–526
Lee S, Kim B, Yang J, Jeong D, Park S, Sang HS, Kook JH, Yang KS, Lee J (2015) Comparative whole genome transcriptome and metabolome analyses of five Klebsiella pneumonia strains. Bioprocess Biosyst Eng 38:2201–2219
Li T, Zhang X, Zhang J, Liu R, Gu C (2018) The transcriptome difference between vulnerable and stable carotid atherosclerotic plaque. Int J Clin Exp Med 11:8988–9004
Markert CL, Shaklee JB, Whitt GS (1975) Evolution of a gene. multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science 189:102–114
Mayer D, Schlensog V, Ck AB (1995) Identification of the transcriptional activator controlling the butanediol fermentation pathway in Klebsiella terrigena. J Bacteriol 177:5261–5269
Nguyen AD, Hwang IY, Lee OK, Kim D, Kalyuzhnaya MG, Mariyana R, Hadiyati S, Kim MS, Lee EY (2018) Systematic metabolic engineering of Methylomicrobium alcaliphilum 20Z for 2,3-butanediol production from methane. Metab Eng 47:323–333
Park JM, Rathnasingh C, Song H (2015) Enhanced production of (R, R)-2,3-butanediol by metabolically engineered Klebsiella oxytoca. J Ind Microbiol Biotechnol 42:1419–1425
Park SW, Lee YJ, Lee WJ, Jee Y, Choi WY (2016) One-step reverse transcription-polymerase chain reaction for ebola and marburg viruses. Int J Env Res Pub Health 7:205–209
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS ONE 7:e30619
Petrov K, Petrova P (2009) High production of 2,3-butanediol from glycerol by Klebsiella pneumoniae G31. Appl Microbiol Biotechnol 84:659–665
Rathnasingh C, Park JM, Kim DK, Song H, Yong KC (2016) Metabolic engineering of Klebsiella pneumoniae and in silico investigation for enhanced 2,3-butanediol production. Biotechnol Lett 38:975–982
Sang JL, Han SC, Chan KK, Thapa LP, Park C, Kim SW (2017) Process strategy for 2,3-butanediol production in fed-batch culture by acetate addition. J Ind Eng Chem 56:157–162
Sikora B, Kubik C, Kalinowska H, Gromek E, Białkowska A, Jędrzejczak-Krzepkowska M, Schüett F, Turkiewicz M (2015) Application of byproducts from food processing for production of 2,3-butanediol using Bacillus amyloliquefaciens TUL 308. Prep Biochem Biotech 46:610–619
Sun LH, Wang XD, Dai JY, **u ZL (2009) Microbial production of 2,3-butanediol from Jerusalem artichoke tubers by Klebsiella pneumoniae. Appl Microbiol Biot 82:847–852
Suwannakham S, Huang Y, Yang ST (2010) Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol Bioeng 94:383–395
Taeyeon K, Sukhyeong C, Sun-Mi L, Min WH, **won L, Youngsoon U, **-Ho S (2016) High production of 2,3-Butanediol (2,3-BD) by Raoultella ornithinolytica B6 via optimizing fermentation conditions and overexpressing 2,3-BD synthesis genes. PLoS ONE 11:0165076
Theissinger K, Falckenhayn C, Blande D, Toljamo A, Gutekunst J, Makkonen J, Jussila J, Lyko F, Schrimpf A, Schulz R, Kokko H (2016) De novo assembly and annotation of the freshwater crayfish Astacus astacus transcriptome. Mar Genomics 28:7–10
Wang P, Zhang J, Feng J, Wang S, Guo L, Wang Y, Lee YY, Taylor S, McDonald T, Wang Y (2019) Enhancement of acid re-assimilation and biosolvent production in Clostridium saccharoperbutylacetonicum through metabolic engineering for efficient biofuel production from lignocellulosic biomass. Bioresource Technol 281:217–225
Yang Z, Zhang Z (2018) Production of (2R, 3R)-2,3-butanediol using engineered Pichia pastoris: strain construction, characterization and fermentation. Biotechnol Biofuels 11:35
Yang T, Rao Z, Zhang X, Xu M, Xu Z, Yang ST (2017) Metabolic engineering strategies for acetoin and 2,3-butanediol production: advances and prospects. Crit Rev Biotechnol 37:1–16
You KO, Park SH, Seol EH, Kim SH, Mi SK, Hwang JW, Dewey DY (2008) Carbon and energy balances of glucose fermentation with hydrogen-producing bacterium Citrobacter amalonaticus Y19. J Microbiol Biotechnol 18:532–538
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant Nos 31570492, 31770544) and Heilongjiang Provincial Key Laboratory of Plant Genetic Engineering and Biological Fermentation Engineering for Cold Region.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ge, J., Wang, J., Ye, G. et al. Disruption of the lactate dehydrogenase and acetate kinase genes in Klebsiella pneumoniae HD79 to enhance 2,3-butanediol production, and related transcriptomics analysis. Biotechnol Lett 42, 537–549 (2020). https://doi.org/10.1007/s10529-020-02802-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02802-7