Abstract
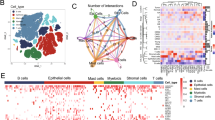
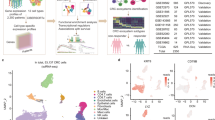
In this study, single-cell RNA-seq data were collected to analyze the characteristics of Histone deacetylation factor (HDF). The tumor microenvironment (TME) cell clusters related to prognosis and immune response were identified by using CRC tissue transcriptome and immunotherapy cohorts from public repository. We explored the expression characteristics of HDF in stromal cells, macrophages, T lymphocytes, and B lymphocytes of the CRC single-cell dataset TME and further identified 4 to 6 cell subclusters using the expression profiles of HDF-associated genes, respectively. The regulatory role of HDF-associated genes on the CRC tumor microenvironment was explored by using single-cell trajectory analysis, and the cellular subtypes identified by biologically characterized genes were compared with those identified by HDF-associated genes. The interaction of HDF-associated gene-mediated microenvironmental cell subtypes and tumor epithelial cells were explored by using intercellular communication analysis, revealing the molecular regulatory mechanism of tumor epithelial cell heterogeneity. Based on the expression of feature genes mediated by HDF-related genes in the microenvironment T-cell subtypes, enrichment scoring was performed on the feature gene expression in the CRC tumor tissue transcriptome dataset. It was found that the feature gene scoring of microenvironment T-cell subtypes (HDF-TME score) has a certain predictive ability for the prognosis and immunotherapy benefits of CRC tumor patients, providing data support for precise immunotherapy in CRC tumors.








Similar content being viewed by others
Availability of Data and Materials
All data are available in a public, open-access repository. The datasets used and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Genome Atlas (TCGA) network (https://cancergenome.nih.gov/). R and other custom scripts for analyzing data are available upon reasonable request.
References
Baidoun F, Elshiwy K, Elkeraie Y et al (2021) Colorectal cancer epidemiology: recent trends and impact on outcomes. Curr Drug Targets 22(9):998–1009
Biffi G, Tuveson DA (2021) Diversity and biology of cancer-associated fibroblasts. Physiol Rev 101(1):147–176
Chen YP, Yin JH, Li WF et al (2020) Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res 30(11):1024–1042
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB (2019) Colorectal cancer. Lancet 394(10207):1467–1480
Gao Y, Wang H, Chen S et al (2022) Single-cell N(6)-methyladenosine regulator patterns guide intercellular communication of tumor microenvironment that contribute to colorectal cancer progression and immunotherapy. J Transl Med 20(1):197
Glozak MA, Seto E (2007) Histone deacetylases and cancer. Oncogene 26(37):5420–5432
Guan XW, Wang HQ, Ban WW et al (2020) Novel HDAC inhibitor chidamide synergizes with Rituximab to inhibit diffuse large B-cell lymphoma tumour growth by upregulating CD20. Cell Death Dis 11(1):20
Hu XT, **ng W, Zhao RS et al (2020) HDAC2 inhibits EMT-mediated cancer metastasis by downregulating the long noncoding RNA H19 in colorectal cancer. J Exp Clin Cancer Res: CR 39(1):270
Li T, Zhang C, Hassan S et al (2018) Histone deacetylase 6 in cancer. J Hematol Oncol 11(1):111
Puram SV, Tirosh I, Parikh AS et al (2017) Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 171(7):1611–1624
Ramaiah MJ, Tangutur AD, Manyam RR (2021) Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci 277:119504
Sung H, Ferlay J, Siegel RL et al (2021) Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin 71(3):209–249
Swierczynski S, Klieser E, Illig R, Alinger-Scharinger B, Kiesslich T, Neureiter D (2015) Histone deacetylation meets miRNA: epigenetics and post-transcriptional regulation in cancer and chronic diseases. Expert Opin Biol Ther 15(5):651–664
von Knethen A, Heinicke U, Weigert A, Zacharowski K, Brüne B (2020) Histone deacetylation inhibitors as modulators of regulatory T cells. Int J Mol Sci 21(7):2356
Acknowledgements
Not applicable
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ZZ—Conceptualization, data curation, writing (original draft). YW—Investigation, software, visualization. XG–Formal analysis, methodology, validation. CY–Formal analysis, methodology, validation. GY—Conceptualization, methodology, writing (review & editing).
Corresponding author
Ethics declarations
Competing interest
The authors have no conflicts of interest to declare.
Ethics Approval and Consent to Participate
Not available.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhao, Z., Wu, Y., Geng, X. et al. Single-Cell Analysis Reveals Histone Deacetylation Factor Guide Intercellular Communication of Tumor Microenvironment that Contribute to Colorectal Cancer Progression and Immunotherapy. Biochem Genet (2024). https://doi.org/10.1007/s10528-024-10730-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-024-10730-8