Abstract
Pseudomonas aeruginosa is one of the top-listed pathogens in nosocomial infection. It is notorious for its complicated virulence system and rapid adaptability to drugs or antimicrobials. In this study, we aimed to evaluate the prevalence of sixteen virulence genes in four groups including type III secretion system, biofilm formation, extracellular toxin biosynthesis and enzymes amongst 209 clinical Pseudomonas aeruginosa strains. We investigated the different distribution patterns of virulence genotypes based on carbapenem-resistant phenotype or the carriage of carbapenemase genes. The detection rate of each virulence gene varied greatly. phzM and plcN were detected in all collected strains, while pilB and exoU were only carried by a small portion of isolates (6.7% and 16.3%). Additionally, the number of genotypes observed in each group of examined virulence genes ranged from 4 to 8. Only the distribution of genotypes of type III secretion system showed statistical difference between carbapenem-mediated or carbapenem-resistant and carbapenem-sensitive strains. The virulence genotype of Pseudomonas aeruginosa was possibly interrelated to its resistance mechanism. Further research suggested that one particular TTSS genotype exhibited higher ratio in carbapenemase-producing strains and exoS was less frequently detected in CRPA strains carrying carbapenemase gene. Generally, the significant genetic diversity of virulence genes amongst Pseudomonas aeruginosa strains was highlighted in this study. Specific TTSS genotypes were associated with carbapenem-resistance. In particular, certain incompatibility might exist between exoS and carbapenemase genes, which provided valuable information for further understanding the relationship between carbapenem resistance and virulence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a common clinical gram-negative bacterium. As one of the top-listed pathogens in nosocomial infections, it is widely distributed on human skin, polluted medical devices and even disinfectants. P. aeruginosa is likely to cause acute or chronic multi-system (e.g., respiratory and urinary tract, bacteremia and soft tissues) infections among immunocompromised individuals (Qin et al. 2022). In addition, the characteristics of rapid adaptability to drugs or antimicrobials resistance and a complicated virulence system make it extremely difficult to treat effectively (Blomquist and Nix 2021).
Pseudomonas aeruginosa can combat the adverse host environment by producing a variety of virulence factors. Its virulence factors are composed of various substances which play an important role in the pathogenic process. Type III secretion system (TTSS) components, encoded by exoU, exoS, exoT and exoY genes, are reported to induce cytotoxic and invasive phenotypes (Galle et al. 2012; Horna and Ruiz 2021). It is one of the most extensively-studied virulence factors with increasing evidence for its predictive value of poor prognosis (Juan et al. 2017). Other extracellular virulence, such as toxins (e.g., toxin A and pyocyanin) and enzymes (e.g., elastases, proteases, phospholipases, and neuraminidases), also contribute to the pathogenicity of bacteria: aiding colonization, destroying protein structures and degrading many crucial proteins of the host cells (Ostroff et al. 1990; Toder et al. 1994; Iiyama et al. 2017). Furthermore, biofilm formation helps P. aeruginosa to survive in hypoxic or other harsh environments, and can lead to chronic infections that are difficult to eradicate (Maurice et al. 2018). Some biofilm matrix molecules (e.g., alginate) and cell appendages (e.g., type IV pili) are necessary for biofilm formation and are also considered as important virulence factors (Mann and Wozniak 2012). Those virulence factors are crucial for P. aeruginosa to cause infection. However, information about prevalence status of these factors amongst clinical isolates in China is inadequate.
In addition to the impact on the pathogenicity of P. aeruginosa, the correlation between virulence and drug resistance has also attracted extensive attention. Many studies had reported that the presence or expression of virulence genes was related to drug resistance. Some studies suggested that they are antagonistic, while other studies reported the resistant P. aeruginosa isolates with high virulence levels (Finlayson and Brown 2011; Jeannot et al. 2008; Pereira et al. 2015). exoU, one remarkable effector of TTSS, was reported to be related to fluoroquinolone resistance based on the results of a higher proportion of fluoroquinolone-resistance among exoU+ isolates (Wong-Beringer et al. 2008; Agnello and Wong-Beringer 2012). Therefore, genes determining virulence and related to drug resistance in P. aeruginosa clinical strains may form a specific combination type through appropriate regulation to better adapt to the hospital environment (Liao et al. 2022). It is meaningful to develop molecular epidemiology researches about virulence-genotype and resistance-phenotype to increase the understanding of P. aeruginosa infection at the molecular or genetic level.
Carbapenem-resistant P. aeruginosa (CRPA) is one of the major challenges in clinical practice, making it more difficult to treat patients (Yoon and Jeong 2021; Kucisec-Tepes 2004). Additionally, it was shown that some CRPA strains may clonally spread worldwide with specific dominant sequence types (Papagiannitsis et al. 2017). This study aimed to evaluate the prevalence of sixteen virulence genes of four virulence types mentioned above (TTSS: exoU, exoS, exoT, exoY; biofilm formation: algD, pilA, pilB; extracellular toxin biosynthesis: toxA, phzM, phzS; enzymes biosynthesis: plcN, plcH, aprA, lasB, nan1 and nan2). We also compared the distribution difference of genotypes amongst subgroups of CRPA and non-CRPA strains. Moreover, this study also investigated whether the presence of particular virulence gene varied with respect to the carriage of carbapenemase gene in CRPA strains.
Materials and methods
Collection and identification of bacterial isolates
209 clinical isolates of P. aeruginosa were collected from 209 distinct clinical patients through eliminating duplicate strains in this study. And they were derived from the Microbiology Department of Laboratory Medicine in Zhongshan Hospital Affiliated to Fudan University in China. The verification of each isolate was based on the growth in blood agar plate at aerobic conditions and the identification was performed by utilizing MALDI-TOF/TOF (bioMérieux, Craponne, France) to single colony with VITEK® MS CHCA (bioMérieux, Craponne, France). MALDI-TOF/TOF achieved the precise microbial identification by its certain proteomic workflows and rigorous machine algorithms. Specifically, unknown bacteria dissolved in matrix solution of CHCA was detected to get the unique protein fingerprint and characteristic peaks which was assigned corresponding weights based on its frequency of occurrence amongst different bacterial strains. The obtained comprehensive information was then compared with the protein fingerprint inside the database. Accordingly, the final identification result and confidence were accurately identified.
Antimicrobial susceptibility test
Antimicrobial susceptibility test of each isolate was detected by Vitek2 Compact system using GN 335 cards (bioMérieux, Craponne, France). It was a miniaturized and simplified version of automatic detection system using microdilution method to determine the minimum inhibitory concentration (MIC). This system was able to achieve automatic dissolution of drugs inside the card using prepared standard bacterial solution. Then, the bacteria growth within the specified time was monitored to obtain the MIC value of each drug. Results were interpreted according to standards established by the Clinical and Laboratory Standards Institute (CLSI. Performance standards for antimicrobial susceptibility testing.). CRPA isolates referred to P. aeruginosa strains that were mediated (MIC: 4 μg/mL) or resistant (MIC: ≥ 8 μg/mL) to imipenem and /or meropenem, Non-CRPA isolates referred to P. aeruginosa strains that were sensitive to imipenem and meropenem (MIC: ≤ 2 μg/mL).
Bacterial genomic DNA isolation
Well-separated colonies of P. aeruginosa strains were cultured in fluid medium of Luria broth (LB) (Solarbio, Shanghai, China) at 37 °C overnight with constant shaking (200 rpm/min). Then, bacterial genomic DNA was isolated by employing the EasyPure® Bacteria Genomic DNA Kit (TransGen Biotech, Bei**g, China) according to the manufacturer’s protocol. DNA samples were then stored at −20 °C before further use in PCR assay for carbapenemase and virulence genes detection.
Carbapenemase and virulence genes detection
Carbapenemase genes (blaKPC, blaGES, blaIMP, blaVIM, blaNDM, blaSIM and blaOXA) that have been reported in China were included in the present study for PCR detection. Sixteen virulence genes noted globally were also examined in this research. Briefly, whole-DNA extracts were detected using 2 × TransTaq®-T PCR SuperMix kit (TransGen Biotech, Bei**g, China) under the following conditions: denaturation for 5 min at 94 °C, followed by 35 cycles of 94 °C for 30 s, 50–60 °C for 30 s, and 72 °C for 30 s, followed by a final extension step of 10 min at 72 °C. The primers involved in this study were listed in Table 1.
PCR product was identified as previously described (Ghanem et al. 2023; Bogiel et al. 2023). It was separated by agarose gel electrophoresis in 1 × Tris–EDTA (TE) buffer. Nucleic Acid Gel Stain (Yeasen Biotechnology, Shanghai, China) achieved the final visual identification of separated bands using gel imaging system (Bio-Rad, Feldkirchen, Germany). P. aeruginosa strains carrying the particular genes and the PAO1 strain were served as positive or negative controls of PCR.
Statistical analysis
All analyses were performed by the software of IBM SPSS Statistics 26.0 (SPPS Inc., Cary, NC). Approach of chi square test (χ2), Fisher-Freeman-Halton test and Criterion of p < 0.05 were used to determine the significance of the observed differences in virulence genes distribution amongst the tested strains, and with respect to their presence into two subgroups of distinct strains (Carbapenemase -positive and Carbapenemase -negative).
Ethics statement
The ethical approval was obtained from the Ethics Committee at Zhongshan Hospital Affiliated to Fudan University (B2022-044R) (Approved on 25 February 2022). All clinical isolates used in this study were collected previously from routine microbiological specimens that were anonymized, and patients were not physically involved in this study.
Results
Clinical isolates
We collected 209 nonrepetitive clinical strains of P. aeruginosa that were comprised of 106 Carbapenem-resistant (CRPA) and 103 Carbapenem-sensitive strains (non-CRPA). As shown in Table S1, they were respectively isolated from respiratory tract (128/209, 61.3%), urine (24/209, 11.5%), drainage (22/209, 10.5%), pus (10/209, 4.8%), bile (9/209, 4.3%), secretion (7/209, 3.3%), hydrothorax and ascites (3, 1.4%), tissue (3/209, 1.4%), catheter (1/209, 0.5%) and other specimens (2/209, 1.0%). Additionally, genes of carbapenemase were positively detected in 20 CRPA strains (20/106, 18.9%).
Prevalence of genes encoding virulence factors
Assessment of examined virulence factors in this study demonstrated a wide variety of gene prevalence. As shown in Fig. 1, the prevalence of four TTSS genes were as follows: exoT (99.52%) > exoY (93.30%) > exoS (78.47%) > exoU (16.27%). And there were fourteen strains carrying these four genes simultaneously.
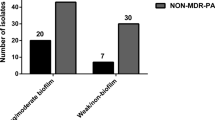
Of the three genes of virulence factors that were related to biofilm biosynthesis (Fig. 2), algD was detected with the highest frequency (84.2%), and pilB with the lowest frequency (6.7%) was the least rare in all examined genes. The detection frequency of pilA was 41.15%. The lone algD was observed in most of clinical isolates, while the number of strains carrying three genes above concurrently was only nine.
The occurrence of the virulence genes involved in producing toxin A and pyocyanin were as follows: toxA-77.51%; phzM-100%; phzS-96.65% (Fig. 3). phzM was detected in all tested strains, and most of them (158) also carried genes of phzS and toxA.
Six enzyme-related virulence genes were detected with generally high frequency in tested strains of this study (Fig. 4). Among which, plcN was noted in all strains, and the detection frequencies of aprA, lasB, nan2 plcH and nan1 were 99.5%, 99.5%, 99.0%, 95.2% and 48.8% respectively. Nearly half of the bacteria strains (94) possessed six genes together.
Virulence genotype and their distributions
As shown in Fig. 5, various genotypes were separately observed in four groups of related virulence genes, and each group exhibited several scarce genotypes.
Genotypes of four groups of virulence genes detected amongst the 209 P. aeruginosa isolates. a, genotypes of TTSS genes amongst the 209 isolates; b, genotypes of genes related to biofilm biosynthesis amongst the 209 isolates; c, genotypes of genes related to the biosynthesis of toxic substances amongst the 209 isolates; d, genotypes of genes related to enzyme production amongst the 209 isolates
Then, a statistically significant difference (Fisher-Freeman-Halton test, p = 0.018) was observed in the overall distributions of TTSS genotypes between CRPA and non-CRPA isolates (Table 2). However, there was no statistical difference in the overall distribution of genotypes in other three groups, which were presented in Table S2, Table S3 and Table S4. Interestingly, TTSS genotype V (exoT+ exoY− exoS− exoU−) isolates were all gathered in CRPA strains although the number of bacterial strains was relatively small (100.0% vs 0.0%, p = 0.002). These results indicated that there might be a specific correlation between particular TTSS genotypes and the phenotype of carbapenem resistance.
The carriage of carbapenemase genes by bacteria is one of the important mechanisms of resistance to carbapenems. To further understand the potential relevance between TTSS genotype and the presence of carbapenemase gene, the distribution of TTSS genotype amongst CRPA strains with carbapenemase gene (carbapenemase +) or not (carbapenemase -) was analysed (Table 3). Specifically, TTSS genotype II exhibited higher ratio in carbapenemase ( +) CRPA (25.0% vs. 4.7%, p = 0.011). Of four genes of TTSS, only exoS was absent in the genotype II, indicating that some certain incompatibility maybe existed between exoS and carbapenemase genes.
Distributions of individual gene of TTSS among CRPA strains
To further investigate the possible association of exoS and carbapenemase gene, we compared the frequency of individual gene of TTSS (exoU, exoS, exoY and exoT) between carbapenemase ( +) and carbapenemase (-) CRPA strains. As shown in Table 4, significant difference in the gene distribution between two subgroups of CRPA strains was observed for exoS (55.00% vs. 88.40%, p = 0.012). exoS was less frequently detected in CRPA strains of carbapenemase ( +), which further implied that it might possess a tendency to be incompatible with genes related to carbapenemase biosynthesis.
Discussion
P. aeruginosa is one of the most important pathogens worldwide characterized by high genetic. The vast majority of isolates possess virulence genes involved in the biosynthesis of pyocyanin, phospholipase C, alkaline protease, elastase B and some exoenzymes. Actually, the virulence trait of P. aeruginosa strain is a complex topic. Particular genotype composition might be one representative facet of their variable pathogenicity potential and sophisticated drug-resistance spectrum, most likely in a subgroup of strains.
The virulence of P. aeruginosa is jointly regulated by multiple signaling layers, and result in the multidimensional harmfulness to the host (Balasubramanian et al. 2013; Jimenez et al. 2012). This study focuses on the epidemiological distribution of important virulence factors related to TTSS, biofilm formation, extracellular toxin and destructive enzyme, and sixteen related genes were evaluated. Specifically, phzM, and plcN were detected in all collected clinical strains. pilB and exoU were only carried by a small portion of strains (6.7% and 16.3%), which is similar to but not entirely consistent with other relevant studies involving clinical isolates from blood or urine samples in Poland and Korea (Park and Koo 2022; Bogiel et al. 2022). Interestingly, one strain of our study carrying all of the sixteen virulence genes was detected. It was also an extensive-drug resistant (XDR) P. aeruginosa with resistance to six types of antibiotics, indicating that there was a possibility that the characteristics of significant drug resistance and high level of virulence could exist simultaneously in the complex medical environment. Besides, an overwhelming majority of P. aeruginosa strains were positively detected for the genes of phzM and phzS that were related to pyocyanin biosynthesis, suggesting that pyocyanin is one of the most common virulence determinants of this species. Fuse et al. had showed that multi-drug resistant (MDR) P. aeruginosa carrying genes coding metallo-β-lactamase (MBL) decreased the pyocyanin-producing ability (Fuse et al. 2013). It was noteworthy that in this study phzM and phzS were identified in only two CRPA isolates producing MBL simultaneously. Additionally, biofilm formation, one of the most common pathogenic factors for P. aeruginosa infection, increased the difficulty of removing the pathogen from the hospital environment and medical equipment (Thi et al. 2020). The detected proportions of related genes of algD (84.21%), pilA (41.15%) and pilB (6.70%) of this study were similar to results reported previously (Bogiel et al. 2022; Kamali et al. 2020; Stewart et al. 2011). Since genes encoding structural pilins of P. aeruginosa were detected at a relatively low frequency, alginate synthesis might be a more significant determinant with regard to the biofilm-associated virulence. In the present study, enzyme-related genes existed in almost all tested isolates except nan1, which was positively detected in about half of P. aeruginosa strains.
Many studies had tried to analyze the potential correlation between virulence and resistance spectrum of P. aeruginosa and got contradictory results. It was previously reported that exoU+ genotype of P. aeruginosa was positively correlated with moderate resistance phenotype and negatively correlated with XDR (Cabot et al. 2012; Peña et al. 2015), and exoU+ genotype was illustrated to be more likely to be resistant to fluoroquinolone with gyrA mutation and overexpression of efflux pump (Wong-Beringer et al. 2008; Agnello and Wong-Beringer 2012). In this study, eight TTSS genotypes of P. aeruginosa were found in 209 clinical isolates, including a rare genotype lacking all the four TTSS genes which was reported previously (Elsen et al. 2014). Additionally, genotypes of TTSS in this study were comprehensively classified based on exoU, exoS, exoY and exoT instead of merely considering exoU like other studies mentioned previously. And TTSS genotype V (exoT+ exoY− exoS− exoU−) of P. aeruginosa with a relatively low level of virulence gene were all resistant to carbapenem, which was likely to support the view of antagonism correlation between virulence and resistance. However, relevant studies were not enough to reveal the association between such genotype and drug resistant pattern. Further research with larger clinical sample size was worthwhile to investigate the association and potential mechanism.
Recently, with increased number of CRPA clinical isolates reported, the situation of carbapenem resistance has become even more severe. CRPA has been listed among the “critical” members of pathogens by WHO, nosocomial infection caused by CRPA and its clonal spread is one of the major global challenges of public health (Daikos et al. 2021; Mancuso et al. 2021). Researches about genetic features, mechanisms of carbapenem resistance and clinical prognosis of CRPA had been widely reported, but studies related to the prevalence of exhaustive virulence genes and their distributions in clinical strains are relatively inadequate. In the present study, the prevalence of the seven genes encoding carbapenemase (blaKPC, blaGES, blaIMP, blaVIM, blaNDM, blaSIM and blaOXA) were also evaluated. The result showed that only 18.9% (20 of 106) of CRPA were positively detected, most of them were blaKPC and blaGES that belong to class A beta-lactamases which have been reported in China (Wang et al. 2006; Ge et al. 2011). Metallo-beta-lactamase (MBL), known as class B beta-lactamases, was most frequently reported in the published data from China (Fang et al. 2022; Feng et al. 2017), while only twol isolates of CRPA were detected that carry blaNDM and blaSIM respectively in this study. Since the studied strains are derived mainly from hospitalized patients in different provinces and cities, indicating that the prevalence of specific genes coding for carbapenemase could vary according to geographical distribution and potential endemic intrahospital spread. The epidemiological characteristic of carbapenemase gene was also reported to be linked to geographical location (Yoon and Jeong 2021). A study showed that the prevalence of carbapenemase gene amongst CRPA strains ranged from 0.0% to 44.3% in multiple global areas (Lee et al. 2022). In the present study, the presence of carbapenemase genes in the CRPA strains accounted for 18.9% of the resistance to carbapenems, which was consistent to the previous study (Zhang et al. 2015). The resistance mechanisms to carbapenems among P. aeruginosa have been found to be diverse. In addition to carrying carbapenemase genes, loss of outer membrane protein OprD and overexpression of efflux pump also contributed to carbapenems resistance (Farra et al. 2008; Masuda et al. 2000; Chalhoub et al. 2016). Therefore, other genetic traits of carbapenem resistance mechanisms mentioned above in the remaining isolates need further investigation.
In this study, we analyzed the different distribution of TTSS genotype based on carbapenem-resistant phenotype or the carriage of carbapenemase genes. It was found that exoS was inclined to reside in carbapenemase (-) CRPA strain, indicating that exoS might be incompatible with genes related to carbapenemase biosynthesis to some extent in clinical strains. We also investigated data from some published researches to analyze the distribution of exoS amongst CRPA strains of Carbapenemase ( +) or Carbapenemase (−) collected from several countries worldwide (Bogiel et al. 2021; Ferreira et al. 2015; Lee et al. 2013; Bellés et al. 2018), Interestingly, the results were consistent to our studies based on clinical isolates of P. aeruginosa from China (Table S5). Since P. aeruginosa had a complicated regulatory network controlling virulence and drug resistance, and the potential antagonism between exoS and carbapenemase gene might be beneficial for bacteria to obtain longer survival and infection (Liang et al. 2014). The finding of specific correlation between some virulence and carbapenemase genes is interesting and worthy of further investigation.
In conclusion, high genetic diversity of virulence was detected amongst clinical P. aeruginosa strains. Many clinical strains were found carrying virulence genes related to the biosynthesis of toxic substances and enzymes, while the prevalence of biofilm-related genes is generally low. Significantly, the distribution of TTSS genotypes showed statistical differences between the subgroup strains of CRPA and non-CRPA, implying the potential correlation between virulence and resistance. Moreover, Significant lower prevalence of exoS was noted in CRPA strains with respect to the presence of carbapenemase gene in this study. This suggested the existence of certain incompatibility between these two genes. It is worthy of further research with larger samples size and in-depth mechanism investigation in the future.
Data availability
Please contact author for data requests.
Abbreviations
- CRPA:
-
Carbapenem-resistant P. aeruginosa
- MBL:
-
Metallo-beta-lactamase
- MDR:
-
Multi-drug resistant
- TTSS:
-
Type III secretion system
- XDR:
-
Extensive-drug resistant
References
Agnello M, Wong-Beringer A (2012) Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS ONE 7(8):e42973. https://doi.org/10.1371/journal.pone.0042973
Balasubramanian D, Schneper L, Kumari H, Mathee K (2013) A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41(1):1–20. https://doi.org/10.1093/nar/gks1039
Bellés A, Bueno J, Rojo-Bezares B, Torres C, Javier Castillo F, Sáenz Y, Seral C (2018) Characterisation of VIM-2-producing Pseudomonas aeruginosa isolates from lower tract respiratory infections in a Spanish hospital. Eur J Clin Microbiol Infect Diseases off Publ Eur Soc Clin Microbiol 37(10):1847–1856. https://doi.org/10.1007/s10096-018-3318-3
Blomquist KC, Nix DE (2021) A Critical evaluation of newer β-lactam antibiotics for treatment of Pseudomonas aeruginosa infections. Ann Pharmacother 55(8):1010–1024. https://doi.org/10.1177/1060028020974003
Bogiel T, Prażyńska M, Kwiecińska-Piróg J, Mikucka A, Gospodarek-Komkowska E (2020) Carbapenem-resistant Pseudomonas aeruginosa strains-distribution of the essential enzymatic virulence factors genes. Antibiotics. https://doi.org/10.3390/antibiotics10010008
Bogiel T, Depka D, Rzepka M, Kwiecińska-Piróg J, Gospodarek-Komkowska E (2021) Prevalence of the genes associated with biofilm and toxins synthesis amongst the Pseudomonas aeruginosa clinical strains. Antibiotics. https://doi.org/10.3390/antibiotics10030241
Bogiel T, Depka D, Rzepka M, Mikucka A (2022) Decoding genetic features and antimicrobial susceptibility of Pseudomonas aeruginosa strains isolated from bloodstream infections. Int J Mol Sci. https://doi.org/10.3390/ijms23169208
Bogiel T, Depka D, Kruszewski S, Rutkowska A, Kanarek P, Rzepka M, Leitão JH, Deptuła A, Gospodarek-Komkowska E (2023) Comparison of virulence-factor-encoding genes and genotype distribution amongst clinical Pseudomonas aeruginosa Strains. Int J Mol Sci. https://doi.org/10.3390/ijms24021269
Cabot G, Ocampo-Sosa AA, Domínguez MA, Gago JF, Juan C, Tubau F, Rodríguez C, Moyà B, Peña C, Martínez-Martínez L et al (2012) Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob Agents Chemother 56(12):6349–6357. https://doi.org/10.1128/aac.01388-12
Chalhoub H, Sáenz Y, Rodriguez-Villalobos H, Denis O, Kahl BC, Tulkens PM, Van Bambeke F (2016) High-level resistance to meropenem in clinical isolates of Pseudomonas aeruginosa in the absence of carbapenemases: role of active efflux and porin alterations. Int J Antimicrob Agents 48(6):740–743. https://doi.org/10.1016/j.ijantimicag.2016.09.012
Daikos GL, da Cunha CA, Rossolini GM, Stone GG, Baillon-Plot N, Tawadrous M, Irani P (2021) Review of Ceftazidime-avibactam for the treatment of infections caused by Pseudomonas aeruginosa. Antibiotics. https://doi.org/10.3390/antibiotics10091126
El-Mahdy R, El-Kannishy G (2019) Virulence factors of Carbapenem-resistant Pseudomonas aeruginosa in hospital-acquired infections in Mansoura. Infection and drug resistance, Egypt. https://doi.org/10.2147/idr.S222329
Elsen S, Huber P, Bouillot S, Couté Y, Fournier P, Dubois Y, Timsit JF, Maurin M, Attrée I (2014) A type III secretion negative clinical strain of Pseudomonas aeruginosa employs a two-partner secreted exolysin to induce hemorrhagic pneumonia. Cell Host Microbe 15(2):164–176. https://doi.org/10.1016/j.chom.2014.01.003
Fang Y, Baloch Z, Zhang W, Hu Y, Zheng R, Song Y, Tai W, **a X (2022) Emergence of carbapenem-resistant ST244, ST292, and ST2446 Pseudomonas aeruginosa clones in burn patients in Yunnan province. Infect Drug Resist. https://doi.org/10.2147/idr.S353130
Farra A, Islam S, Strålfors A, Sörberg M, Wretlind B (2008) Role of outer membrane protein OprD and penicillin-binding proteins in resistance of Pseudomonas aeruginosa to imipenem and meropenem. Int J Antimicrob Agents 31(5):427–433. https://doi.org/10.1016/j.ijantimicag.2007.12.016
Feng W, Sun F, Wang Q, **ong W, Qiu X, Dai X, **a P (2017) Epidemiology and resistance characteristics of Pseudomonas aeruginosa isolates from the respiratory department of a hospital in China. J Glob Antimicrob Resist. https://doi.org/10.1016/j.jgar.2016.11.012
Ferreira ML, Dantas RC, Faria ALS, Gonçalves IR, Silveira de Brito C, Queiroz LL, Gontijo-Filho PP, Ribas RM (2015) Molecular epidemiological survey of the quinolone- and carbapenem-resistant genotype and its association with the type III secretion system in Pseudomonas aeruginosa. J Med Microbiol 64(Pt 3):262–271. https://doi.org/10.1099/jmm.0.000023
Finlayson EA, Brown PD (2011) Comparison of antibiotic resistance and virulence factors in pigmented and non-pigmented Pseudomonas aeruginosa. West Indian Med J 60(1):24–32
Fuse K, Fujimura S, Kikuchi T, Gomi K, Iida Y, Nukiwa T, Watanabe A (2013) Reduction of virulence factor pyocyanin production in multidrug-resistant Pseudomonas aeruginosa. J Infect Chemother off J Japan Soc Chemother 19(1):82–88. https://doi.org/10.1007/s10156-012-0457-9
Galle M, Carpentier I, Beyaert R (2012) Structure and function of the Type III secretion system of Pseudomonas aeruginosa. Curr Protein Pept Sci 13(8):831–842. https://doi.org/10.2174/138920312804871210
Ge C, Wei Z, Jiang Y, Shen P, Yu Y, Li L (2011) Identification of KPC-2-producing Pseudomonas aeruginosa isolates in China. J Antimicrob Chemother 66(5):1184–1186. https://doi.org/10.1093/jac/dkr060
Ghanem SM, Abd El-Baky RM, Abourehab MAS, Fadl GFM, Gamil N (2023) Prevalence of quorum sensing and virulence factor genes among Pseudomonas aeruginosa isolated from patients suffering from different infections and their association with antimicrobial resistance. Infect Drug Resist. https://doi.org/10.2147/idr.S403441
Hayashi W, Izumi K, Yoshida S, Takizawa S, Sakaguchi K, Iyori K, Minoshima KI, Takano S, Kitagawa M, Nagano Y et al (2021) Antimicrobial resistance and type III secretion system Virulotypes of Pseudomonas aeruginosa isolates from dogs and cats in primary veterinary hospitals in Japan: identification of the international high-risk clone sequence type 235. Microbiol Spect 9(2):e0040821. https://doi.org/10.1128/Spectrum.00408-21
Horna G, Ruiz J (2021) Type 3 secretion system of Pseudomonas aeruginosa. Microbiol Res. https://doi.org/10.1016/j.micres.2021.126719
Iiyama K, Takahashi E, Lee JM, Mon H, Morishita M, Kusakabe T, Yasunaga-Aoki C (2017) Alkaline protease contributes to pyocyanin production in Pseudomonas aeruginosa. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fnx051
Jeannot K, Elsen S, Köhler T, Attree I, van Delden C, Plésiat P (2008) Resistance and virulence of Pseudomonas aeruginosa clinical strains overproducing the MexCD-OprJ efflux pump. Antimicrob Agents Chemother 52(7):2455–2462. https://doi.org/10.1128/aac.01107-07
Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76(1):46–65. https://doi.org/10.1128/mmbr.05007-11
Juan C, Peña C, Oliver A (2017) Host and pathogen biomarkers for severe Pseudomonas aeruginosa infections. J Infect Diseases 215:S44-s51. https://doi.org/10.1093/infdis/jiw299
Kamali E, Jamali A, Ardebili A, Ezadi F, Mohebbi A (2020) Evaluation of antimicrobial resistance, biofilm forming potential, and the presence of biofilm-related genes among clinical isolates of Pseudomonas aeruginosa. BMC Res Notes 13(1):27. https://doi.org/10.1186/s13104-020-4890-z
Kucisec-Tepes N (2004) Pseudomonas aeruginosa–a significant hospital pathogen and resistance to carbapenem. Acta Medica Croatica : Casopis Hravatske Akademije Medicinskih Znanosti 58(4):313–321
Lee JY, Peck KR, Ko KS (2013) Selective advantages of two major clones of carbapenem-resistant Pseudomonas aeruginosa isolates (CC235 and CC641) from Korea: antimicrobial resistance, virulence and biofilm-forming activity. J Med Microbiol 62(Pt 7):1015–1024. https://doi.org/10.1099/jmm.0.055426-0
Lee YL, Ko WC, Hsueh PR (2022) Geographic patterns of carbapenem-resistant pseudomonas aeruginosa in the Asia-Pacific Region: results from the antimicrobial testing leadership and surveillance (atlas) program, 2015–2019. Antimicrob Agents Chemother 66(2):e0200021. https://doi.org/10.1128/aac.02000-21
Liang H, Deng X, Li X, Ye Y, Wu M (2014) Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res 42(16):10307–10320. https://doi.org/10.1093/nar/gku586
Liao C, Huang X, Wang Q, Yao D, Lu W (2022) Virulence factors of Pseudomonas aeruginosa and antivirulence strategies to combat its drug resistance. Front Cell Infect Microbiol. https://doi.org/10.3389/fcimb.2022.926758
Mancuso G, Midiri A, Gerace E, Biondo C (2021) Bacterial antibiotic resistance: the most critical pathogens. Pathogens. https://doi.org/10.3390/pathogens10101310
Mann EE, Wozniak DJ (2012) Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol Rev 36(4):893–916. https://doi.org/10.1111/j.1574-6976.2011.00322.x
Masuda N, Sakagawa E, Ohya S, Gotoh N, Tsujimoto H, Nishino T (2000) Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother 44(12):3322–3327. https://doi.org/10.1128/aac.44.12.3322-3327.2000
Maurice NM, Bedi B, Sadikot RT (2018) Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am J Respir Cell Mol Biol 58(4):428–439. https://doi.org/10.1165/rcmb.2017-0321TR
Ostroff RM, Vasil AI, Vasil ML (1990) Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J Bacteriol 172(10):5915–5923. https://doi.org/10.1128/jb.172.10.5915-5923.1990
Papagiannitsis CC, Medvecky M, Chudejova K, Skalova A, Rotova V, Spanelova P, Jakubu V, Zemlickova H, Hrabak J (2017) Molecular characterization of Carbapenemase-producing Pseudomonas aeruginosa of Czech Origin and evidence for clonal spread of extensively resistant sequence type 357 expressing IMP-7 metallo-β-lactamase. Antimicrob Agents Chemother. https://doi.org/10.1128/aac.01811-17
Park Y, Koo SH (2022) Epidemiology, molecular characteristics, and virulence factors of Carbapenem-resistant Pseudomonas aeruginosa isolated from patients with urinary tract infections. Infect Drug Resist. https://doi.org/10.2147/idr.S346313
Peña C, Cabot G, Gómez-Zorrilla S, Zamorano L, Ocampo-Sosa A, Murillas J, Almirante B, Pomar V, Aguilar M, Granados A et al (2015) Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin Infect Diseases off Publ Infect Diseases Soc Am 60(4):539–548. https://doi.org/10.1093/cid/ciu866
Pereira SG, Rosa AC, Cardoso O (2015) Virulence factors as predictive tools for drug resistance in Pseudomonas aeruginosa. Virulence 6(7):679–683. https://doi.org/10.1080/21505594.2015.1048958
Petit SM, Lavenir R, Colinon-Dupuich C, Boukerb AM, Cholley P, Bertrand X, Freney J, Doléans-Jordheim A, Nazaret S, Laurent F et al (2013) Lagooning of wastewaters favors dissemination of clinically relevant Pseudomonas aeruginosa. Res Microbiol 164(8):856–866. https://doi.org/10.1016/j.resmic.2013.06.007
Poirel L, Walsh TR, Cuvillier V, Nordmann P (2011) Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis 70(1):119–123. https://doi.org/10.1016/j.diagmicrobio.2010.12.002
Qin S, **ao W, Zhou C, Pu Q, Deng X, Lan L, Liang H, Song X, Wu M (2022) Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct Target Ther 7(1):199. https://doi.org/10.1038/s41392-022-01056-1
Queenan AM, Bush K (2007) Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20(3):440–458. https://doi.org/10.1128/cmr.00001-07
Stewart RM, Wiehlmann L, Ashelford KE, Preston SJ, Frimmersdorf E, Campbell BJ, Neal TJ, Hall N, Tuft S, Kaye SB et al (2011) Genetic characterization indicates that a specific subpopulation of Pseudomonas aeruginosa is associated with keratitis infections. J Clin Microbiol 49(3):993–1003. https://doi.org/10.1128/jcm.02036-10
Thi MTT, Wibowo D, Rehm BHA (2020) Pseudomonas aeruginosa Biofilms. Int J Mol Sci. https://doi.org/10.3390/ijms21228671
Toder DS, Ferrell SJ, Nezezon JL, Rust L, Iglewski BH (1994) lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect Immun 62(4):1320–1327. https://doi.org/10.1128/iai.62.4.1320-1327.1994
Wang C, Cai P, Chang D, Mi Z (2006) A Pseudomonas aeruginosa isolate producing the GES-5 extended-spectrum beta-lactamase. J Antimicrob Chemother 57(6):1261–1262. https://doi.org/10.1093/jac/dkl116
Wong-Beringer A, Wiener-Kronish J, Lynch S, Flanagan J (2008) Comparison of type III secretion system virulence among fluoroquinolone-susceptible and -resistant clinical isolates of Pseudomonas aeruginosa. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Diseases 14(4):330–336. https://doi.org/10.1111/j.1469-0691.2007.01939.x
Yoon EJ, Jeong SH (2021) Mobile carbapenemase genes in Pseudomonas aeruginosa. Front Microbiol. https://doi.org/10.3389/fmicb.2021.614058
Zhang X, Gu B, Mei Y, Wen Y, **a W (2015) Increasing resistance rate to carbapenem among blood culture isolates of Klebsiella pneumoniae, Acinetobacter baumannii and Pseudomonas aeruginosa in a university-affiliated hospital in China, 2004–2011. J Antibiot 68(2):115–120. https://doi.org/10.1038/ja.2014.119
Funding
This work was supported by National Natural Science Foundation of China (82130001), Science and Technology Commission of Shanghai Municipality (20Z11901000, 20DZ2261200, 20XD1401200), Shanghai Municipal Key Clinical Specialty (shslczdzk02201), Shanghai Municipal Health Commission and Shanghai Municipal Administrator of Traditional Chinese Medicine (ZY(2021-2023)-0207-01).
Author information
Contributions
YS, WG, XW contributed to the study conception and design. Material preparation was performed by XW, KG, YS, CC, CZ and CZ. Data collection was performed by XW and KG. Analysis was performed by XW and KG. The draft of the manuscript was written by XW. And all authors commented on previous versions of the manuscript. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that they have no conflicts of interest.
Consent for publication
Not Applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, X., Gao, K., Chen, C. et al. Prevalence of the virulence genes and their correlation with carbapenem resistance amongst the Pseudomonas aeruginosa strains isolated from a tertiary hospital in China. Antonie van Leeuwenhoek 116, 1395–1406 (2023). https://doi.org/10.1007/s10482-023-01869-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-023-01869-2