Abstract
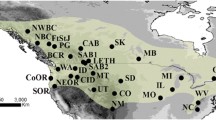
For more than half a century, the Western Eurasian Crane (Grus grus grus) has been expanding its range toward western Europe, recolonizing areas where it had been previously driven to extinction, including the UK, the Netherlands and Denmark. The Western Eurasian Crane is, on the one hand, a very mobile, migratory species, but on the other, is territorial and shows high breeding site fidelity. Hence, its genetic population structure is subject to antagonizing forces, which have different consequences. Based on the genoty** of six highly variable microsatellite loci, we inferred the population structure of the Western Eurasian Crane from samples from eight regions. We integrated classic F-statistics including analyses of molecular variance with a priori designation of structure and divisive clustering approaches, i.e. a Bayesian procedure (STRUCTURE) and discriminant analysis of principal components, which infer structure a posteriori. According to the F-statistics, populations were only weakly differentiated, and the majority of the genetic variance (> 90%) was attributed to individuals. At first glance, the divisive approaches appeared to agree in finding four clusters. Yet, there was no correspondence regarding the composition of the clusters and none of the results were biologically meaningful. However, STRUCTURE delivered an alternative interpretation, designating the highest likelihood to a scenario without subdivision, in clear agreement with the findings based on the F-statistics. In conclusion, the Western Eurasian Crane is genetically largely homogeneous, probably as a consequence of the rapid growth and range expansion of its population.
Zusammenfassung
Geringe genetische Populationsstruktur eines sich ausbreitenden Zugvogels mit hoher Brutortreue, des Westlichen Eurasischen Kranichs, Grus grus grus
Seit mehr als einem halben Jahrhundert breitet sich der Eurasische Kranich (Grus grus grus) wieder nach Westeuropa bis hin nach Großbritannien, den Niederlanden und Dänemark aus, wo er ausgerottet war. Der Eurasische Kranich ist einerseits eine sehr mobile, ziehende Art. Andererseits ist er territorial und zeichnet sich durch eine hohe Brutplatztreue aus. Die genetische Populationsstruktur ist somit von gegensätzlichen Kräften mit unterschiedlichen Konsequenzen geprägt. Wir ermittelten die Populationsstruktur der Westeuropäischen Population (WEP) des Eurasischen Kranichs basierend auf sechs hochvariablen Mikrosatelliten-Loci und Samples aus acht Regionen. Wir kombinierten klassische F-Statistik einschließlich molekularer Varianzanzanalysen mit a priori festgelegter Struktur mit divisiven Clusteranalysen—einer Bayes’schen Methode (STRUCTURE) und einer Diskriminanzanalyse von Hauptkomponenten (DAPC)–, die die Struktur a posteriori schätzen. Die F-Statistik zeigte, dass die Populationen nur gering differenziert waren. Der Großteil der genetischen Varianz (> 90%) lag auf Ebene der Individuen. Auf den ersten Blick schienen die divisiven Ansätze ein übereinstimmendes Bild zu zeichnen. Beide fanden vier Cluster. Allerdings gab es keinerlei Übereinstimmung in der Zusammensetzung der Cluster und keines der Resultate war biologisch sinnvoll. STRUCTURE wies allerdings die höchste Wahrscheinlichkeit einem Szenario ohne Populationsunterteilung zu und lieferte somit eine alternative Interpretation, die mit der F-Statistik übereinstimmte. Daher schließen wir, dass die WEP des Eurasischen Kranichs weitestgehend homogen ist.



Similar content being viewed by others
References
Barton N, Slatkin M (1986) A quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56:409–415
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Cons Gen Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L (2004) Patterns of DNA sequence diversity and genetic structure after a range expansion: lessons from the infinite-island model. Mol Ecol 13:853–864
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under linux and windows. Mol Ecol Resour 10:564–567
Excoffier L, Foll M, Petit RJ (2009) Genetic consequences of range expansions. Annu Rev Ecol Evol Syst 40:481–501
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fourcade Y, Richardson DS, Keišs O, Budka M, Green RE, Fokin S, Secondi J (2016) Corncrake conservation genetics at a European scale: the impact of biogeographical and anthropological processes. Biol Cons 198:210–219
Garnier J, Lewis MA (2016) Expansion under climate change: the genetic consequences. Bull Math Biol 78:2165–2185
Goldberg J, Trewick SA, Powlesland RG (2011) Population structure and biogeography of Hemiphaga pigeons (Aves: Columbidae) on islands in the New Zealand region. J Biogeogr 38:285–298
Goodsman DW, Cooke B, Coltman DW, Lewis MA (2014) The genetic signature of rapid range expansions: how dispersal, growth and invasion speed impact heterozygosity and allele surfing. Theor Pop Biol 98:1–10
Haase M, Ilyashenko V (2012) A glimpse on mitochondrial differentiation of four currently recognized subspecies of Common Crane (Grus grus). Ardeola 59:131–136
Hewitt G (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Höltje H, Mewes W, Haase M, Schmitz A (2016) Genetic evidence of female specific eggshell colouration in the Common Crane (Grus grus). J Ornithol 157:609–617
Horváth MBB, Martinez-Cruz JJ, Negro L, Kalm R, Godoy JA (2005) An overlooked DNA source for non-invasive genetic analysis in birds. J Avian Biol 36:84–88
Ibrahim KM, Nichols RA, Hewitt GM (1996) Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 77:282–291
Jahner JP, Gibson D, Weitzman CL, Blomberg EJ, Selinger JS, Parchman TL (2016) Fine-scale genetic structure among Greater Sage-grouse leks in central Nevada. BMC Evol Biol 16:127
Jombart T, Ahmed I (2011) Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Joosten H, Couwenberg J (2001) Zur Anthropogenen Veränderung der Moore: Bilanzen zum Moorverslust, das Beispiel Europa. In: Succow M, Joosten H (eds) Landschaftsökologische Moorkunde. Schweizerbart’sche, Stuttgart, pp 406–409
Jost L (2008) G ST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191
Leito A, Keskpaik J, Ojaste I, Truu J (2006) The Eurasian Crane in Estonia. Estonian University of Life Sciences, Tartu
Leito A, Ojaste I, Põder I (2013) Monitoring of the Eurasian Crane in Estonia: methods and last results. In: Nowald G, Weber A, Fanke J, Weinhardt E, Donner N (eds) Proceedings of the VIIth European Crane Conference, Stralsund. Groß Mohrdorf, pp 141–145
Leito A, Külvik M, Bunce RGH, Ojaste I, Raet J, Villoslada M, Leivits M, Kull A, Kuusemets V, Kull T, Metzger MJ, Sepp K (2015) The potential impacts of changes in ecological networks, land use and climate on the Eurasian Crane population in Estonia. Landscape Ecol 30:887–904
Meirmans PG, Hedrick PW (2011) Assessing population structure: F ST and related measures. Mol Ecol Resour 11:5–18
Meirmans PG, van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Mewes W (2017) Die Brutorttreue von Kranichen Grus grus in Nordostdeutschland. Vogelwelt 137:249–260
Mills LS, Allendorf FW (1996) The one-migrant-per-generation-rule in conservation and management. Cons Biol 6:1509–1518
Morinha F, Dávila JA, Bastos E, Cabral JA, Frías O, Gonzalez JL, Travassos P, Carvalho D, Milá B, Blanco G (2017) Extreme genetic structure in a social bird species despite high dispersal capacity. Mol Ecol 26:2812–2825
Mudrik EA, Kashentseva TA, Redchuk PS, Politov DV (2015) Microsatellite variability data confirm low genetic differentiation of western and eastern subspecies of Common Crane Grus grus L. (Gruidae, Aves). Mol Biol 49:260–266
Nowald G, Alonso JC (2016) Beringung und Datenverarbeitung. In: Prange H (ed) Die Welt der Kraniche. MediaNatur, Minden, pp 183–194
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research: an update. Bioinformatics 28:2537–2539
Prange H (2016) Die Welt der Kraniche. MediaNatur, Minden
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for STRUCTURE software: version 2.2
Proctor NS, Lynch PJ (1993) Manual of ornithology: avian structure and function. Yale University Press, Yale
Pulgarín-R PC, Burg TM (2012) Genetic signals of demographic expansion in Downy Woodpecker (Picoides pubescens) after the last North American Glacial Maximum. PLOS ONE 7:e40412
Quillfeldt P, Moodley Y, Weimerskirch H, Cherel Y, Delord K, Phillips RA, Navarro J, Calderon L, Masello JF (2017) Does genetic structure reflect differences in non-breeding movements? A case study in small, highly mobile seabirds. BMC Evol Biol 17:160
R Development Core Team (2013) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Ross KG (2001) Molecular ecology of social behaviour: analyses of breeding systems and genetic structure. Mol Ecol 10:263–284
Rousset F (2007) Inferences from spatial population genetics. In: Balding D, Bishop M, Cannings C (eds) Handbook of statistical genetics. Wiley, Chichester, pp 945–979
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Storfer A, Murphy MA, Evans JS, Goldberg CS, Robinson S, Spear SF, Dezzani R, Delmelle E, Vierling L, Waits LP (2007) Putting the “landscape” in landscape genetics. Heredity 98:128–142
Tofft J (2016) Dänemark. In: Prange H (ed) Die Welt der Kraniche. MediaNatur, Minden, pp 462–465
Van Andel J, Aronson J (2006) Restoration ecology. Blackwell, Malden
Wang J (2004) Application of the one-migrant-per-generation rule to conservation and management. Cons Biol 18:332–343
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: F ST not equal to 1/(4N m + 1). Heredity 82:117–125
Wright S (1978) Evolution and the genetics of populations. Variability within and among natural populations, vol 4. University of Chicago Press, Chicago
Zaccara S, Crosa G, Vanetti I, Binelli G, Childress B, McCulloch G, Harper DM (2011) Lesser Flamingo Phoeniconaias minor as a nomadic species in African shallow alkaline lakes and pans: genetic structure and future perspectives. Ostrich 82:95–100
Acknowledgements
Completely unexpectedly, our co-author Aivar Leito passed away a few days before finalizing the manuscript. We dedicate this study to his memory. We are grateful to Christel Meibauer and Silke Fregin, technicians in the lab in Greifswald, for their support. Pekka Mustakallio is particularly acknowledged for organising the Finnish crane ringing scheme. We are thankful to Uko Bleive, Trinus Haitjema, Mati Martinson, Indrek Põder, Urmas Sellis and Ainar Unus for collecting the samples in Estonia. This work was supported by institutional research funding (IUT21-1) of the Estonian Ministry of Education and Research. E. A. M. and D. V. P. were supported by the Russian state (contract no. 0112-2019-0001).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wink.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Haase, M., Höltje, H., Blahy, B. et al. Shallow genetic population structure in an expanding migratory bird with high breeding site fidelity, the Western Eurasian Crane Grus grus grus. J Ornithol 160, 965–972 (2019). https://doi.org/10.1007/s10336-019-01688-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-019-01688-1