Abstract
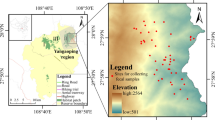
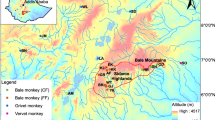
The endemic Samango monkey subspecies (Cercopithecus albogularis labiatus) inhabits small discontinuous Afromontane forest patches in the Eastern Cape, KwaZulu-Natal midlands and southern Mpumalanga Provinces in South Africa. The subspecies is affected by restricted migration between forest patches which may impact on gene flow resulting in inbreeding and possible localized extinction. Current consensus, based on habitat quality, is that C. a. labiatus can be considered as endangered as the small forest patches they inhabit may not be large enough to sustain them. The aim of this study was to conduct a molecular genetic investigation to determine if the observed isolation has affected the genetic variability of the subspecies. A total of 65 Samango monkeys (including juveniles, subadults and adults) were sampled from two localities within the Hogsback area in the Amathole Mountains. Nuclear and mitochondrial DNA variation was assessed using 17 microsatellite markers and by sequencing the hypervariable 1 region (HVR1). Microsatellite data generated was used to determine population structure, genetic diversity and the extent of inbreeding. Sequences of the HVR1 were used to infer individual origins, haplotype sharing and haplotype diversity. No negative genetic factors associated with isolation such as inbreeding were detected in the two groups and gene flow between groups can be regarded as fairly high primarily as a result of male migration. This was in contrast to the low nuclear genetic diversity observed (H o = 0.45). A further reduction in heterozygosity may lead to inbreeding and reduced offspring fitness. Translocations and establishment of habitat corridors between forest patches are some of the recommendations that have emerged from this study which will increase long-term population viability of the subspecies.







Similar content being viewed by others
References
Allen JM, Miyamoto MM, Wu CH, Carter TE, Ungvari-Martin J, Magrini K, Chapman CA (2012) Primate DNA suggests long-term stability of an African rainforest. Ecol Evol 2:2829–2842
Booth CP (1968) Taxonomic studies of Cercopithecus mitis wolf (East Africa). Nat Geog Soc Res Reports (1963 projects):37–51
Bowler DE, Benton TG (2005) Causes and consequences of animal dispersal strategies: relating individual behaviour to spatial dynamics. Biol Rev 80:205–225
Butynski TM (1990) Comparative ecology of blue monkeys (Cercopithecus mitis) in high-and low-density subpopulations. Ecol Monogr 60:1–26
Butynski TM (2002) Conservation of the guenons: an overview of status, threats, and recommendations. In: Glenn ME, Cords M (eds) The guenons: diversity and adaptation in African monkeys. Developments in primatology: progress and prospects . Springer, Boston, pp 411–424
Carlson AA, Isbell LA (2001) Causes and consequences of single-male and multimale mating in free-ranging patas monkeys, Erythrocebus patas. Anim Behav 62:1047–1058
Chang Z, Yang B, Vigilant L, Liu Z, Ren B, Yang J, **ang Z, Garber PA, Li M (2014) Evidence of male-biased dispersal in the endangered Sichuan snub-nosed monkey (Rhinopithexus roxellana). Am J Primatol 76:72–83
Colyn M (1991) Zoogeographical importance of Zaire River basin for speciation (translated). Annalen der koninklijk Museum voor Midden-Afrika, Zool Wetenschappen, Tervuren, Belgium 264:1–250
Dalton DL, Linden B, Wimberger K, Nupen LJ, Tordiffe AS, Taylor PJ, Madisha MT, Kotze A (2015) New insights into Samango monkey speciation in South Africa. PLoS One 10:e0117003
Earl DA, Vonholdt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Ekernas L, Cords M (2007) Social and environmental factors influencing natal dispersal in blue monkeys, Cercopithecus mitis stuhlmanni. Anim Behav 73:1009–1020
Ely B, Wilson JL, Jackson F, Jackson BA (2006) African-American mitochondrial DNAs often match mtDNAs found in multiple African ethnic groups. BMC Biol 4:1
Enstam KL, Isbell LA (2007) The guenons (Genus: Cercopithecus) and their allies: behavioral ecology of polyspecific associations. In: Campbell CJ, Fuentes A, MacKinnon KC, Panger MA, Bearder SK (eds) Primates in perspective, 1st edn. Oxford University Press, Oxford, pp 252–273
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Everard DA, Hardy SD (1993) Composition, structure and dynamics of the Amatola forests. Report FOR-DEA 810. Division of Forest Science and Technology, CSIR, Pretoria
Fahrig L (2003) Effects of habitat fragmentation on biodiversity. Annu Rev Ecol Evol Syst 34:487–515
Fischer J, Lindenmayer DB (2007) Landscape modification and habitat fragmentation: a synthesis. Global Ecol Biogeogr 16:265–280
Friedman Y, Daly B (2004) Red data book of the mammals of South Africa: a conservation assessment: conservation breeding specialist group southern Africa. Conservation Breeding Specialist Group (SSC/IUCN), Endangered Wildlife Trust, Saxonwold, South Africa
Gautier-Hion A, Colyn M, Gautier JP (1999) Natural history of Central African primates. Ecofac, Libreville, pp 43–46
Goossens B, Chikhi L, Jalil MF, Ancrenaz M, Lackman-Ancrenaz I, Mohamed M, Andau P, Bruford MW (2005) Patterns of genetic diversity and migration in increasingly fragmented and declining orang-utan (Pongo pygmaeus) populations from Sabah, Malaysia. Mol Ecol 14:441–456
Griffith B, Scott JM, Carpenter JW, Reed C (1989) Translocation as a species conservation tool: status and strategy. Science 245:477–480
Groves CP (2001) Primate taxonomy. Smithsonian Institution Press, Washington
Groves CP (2005) Geographic variation within eastern chimpanzees (Pan troglodytes cf. schweinfurthii Giglioli, 1872). Australas Primatol 17:19–46
Grubb P, Butynski TM, Oates JF, Bearder SK, Disotell TR, Groves CP, Struhsaker TT (2003) Assessment of the diversity of African primates. Int J Primatol 24:1301–1357
Guschanski K, Krause J, Sawyer S, Valente LM, Bailey S, Finstermeier K, Sabin R, Gilissen E, Sonet G, Nagy ZT, Lenglet G, Mayer F, Savolainen V (2013) Next-generation museomics disentangles one of the largest primate radiations. Syst Biol 62:539–554
Haus T, Roos C, Zinner D (2013) Discordance between spatial distributions of Y-chromosomal and mitochondrial haplotypes in African green monkeys (Chlorocebus spp.): a result of introgressive hybridization or cryptic diversity? Int J Primatol 34:986–999
Heyns H, Ferguson, M, Pitcford W, Seydack A (1989) Bestuursplan: Oos-kaap woude. Unpublished report
Lawes MJ (1990) The distribution of the Samango monkey (Cercopithecus mitis erythrarchus peters, 1852 and Cercopithecus mitis labiatus, Geoffroy, 1843) and forest history in southern Africa. J Biogeogr 17:669–680
Lebigre C, Alatalo RV, Siitari H (2010) Female-biased dispersal alone can reduce the occurrence of inbreeding in black grouse (Tetrao tetrix). Mol Ecol 19:1929–1939
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Low AB, Rebelo AG (1996) Vegetation of South Africa. Lesotho and Swaziland, Pretoria, p p27
Melnick DJ, Hoelzer GA (1992) Differences in male and female macaque dispersal lead to contrasting distributions of nuclear and mitochondrial DNA variation. Int J Primatol 13:379–393
Meyer D, Rinaldi ID, Ramlee H, Perwitasari-Farajallah D, Hodges JK, Roos C (2011) Mitochondrial phylogeny of leaf monkeys (gensu Presbytis, Eschscholtz, 1821) with implications for taxonomy and conservation. Mol Phylogenet Evol 59:311–319
Napier PH (1981) Catalogue of primates in the British Museum (Natural History) and elsewhere in the British Isles. Part II: family Cercopithecidae, subfamily Cercopithecinae. British Museum (Natural History), London
O’Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R (2006) Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv 133:42–51
Oklander LI, Kowalewski MM, Corach D (2010) Genetic consequences of habitat fragmentation in black-and-gold howler (Alouatta caraya) populations from Northern Argentina. Int J Primatol 31:813–832
Park S (2001) Microsatellite toolkit. Department of Genetics, Trinity College, Dublin
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Prugnolle F, De Meeûs T (2002) Inferring sex-biased dispersal from population genetic tools: a review. Heredity 88:161–165
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reed DH, Briscoe DA, Frankham R (2002) Inbreeding and extinction: effects of environmental stress and lineages. Conserv Genet 3:301–307
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–569
Skinner JD, Chimimba CT (2005) The mammals of the southern African sub-region. Cambridge University Press, Cambridge
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Toews DPL, Brelsford A (2012) The biogeography of mitochondrial and nuclear discordance in animals. Mol Ecol 21:3907–3930
Van Oosterhout C, Hutchinson B, Wills D, Shipley P (2004) Micro-checker: software for identifying and correcting genoty** errors in microsatellite data. Mol Ecol Notes 4:535–538
White F (1983) The vegetation of Africa. United Nations Scientific and Cultural Organization, Paris
Whitfield LS, Lovell-Badge R, Goodfellow PN (1993) Rapid sequence evolution of the mammalian sex-determining gene SRY. Nature 364:713–715
Acknowledgements
We thank Dr. Kirsten Wimberger and Dr. Adrian S.W Tordiffe for fieldwork conducted to obtain samples used in this study. We acknowledge the National Research Foundation (NRF) and the National Zoological Gardens of South Africa (NZG) for organizational core grant funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical considerations
Ethical approval for this study was obtained from the Research Ethics & Scientific Committee (RESC, Project #2012/P12/08) of the National Zoological Gardens of South Africa (NZG) and by the Tshwane University of Technology’s (TUT) Animal Research Ethics Committee (Reference Number #AREC2014/06/001). All samples used in this project were stored at the National Zoological Gardens Biobank in Pretoria.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Madisha, M.T., Dalton, D.L., Jansen, R. et al. Genetic assessment of an isolated endemic Samango monkey (Cercopithecus albogularis labiatus) population in the Amathole Mountains, Eastern Cape Province, South Africa. Primates 59, 197–207 (2018). https://doi.org/10.1007/s10329-017-0636-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10329-017-0636-5