Abstract
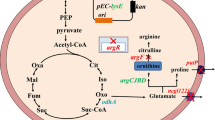
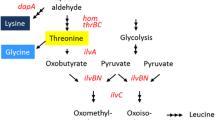
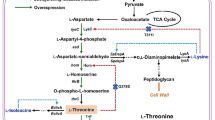
Previously we have characterized a threonine dehydratase mutant TDF383V (encoded by ilvA1) and an acetohydroxy acid synthase mutant AHASP176S, D426E, L575W (encoded by ilvBN1) in Corynebacterium glutamicum IWJ001, one of the best l-isoleucine producing strains. Here, we further characterized an aspartate kinase mutant AKA279T (encoded by lysC1) and a homoserine dehydrogenase mutant HDG378S (encoded by hom1) in IWJ001, and analyzed the consequences of all these mutant enzymes on amino acids production in the wild type background. In vitro enzyme tests confirmed that AKA279T is completely resistant to feed-back inhibition by l-threonine and l-lysine, and that HDG378S is partially resistant to l-threonine with the half maximal inhibitory concentration between 12 and 14 mM. In C. glutamicum ATCC13869, expressing lysC1 alone led to exclusive l-lysine accumulation, co-expressing hom1 and thrB1 with lysC1 shifted partial carbon flux from l-lysine (decreased by 50.1 %) to l-threonine (4.85 g/L) with minor l-isoleucine and no l-homoserine accumulation, further co-expressing ilvA1 completely depleted l-threonine and strongly shifted carbon flux from l-lysine (decreased by 83.0 %) to l-isoleucine (3.53 g/L). The results demonstrated the strongly feed-back resistant TDF383V might be the main driving force for l-isoleucine over-synthesis in this case, and the partially feed-back resistant HDG378S might prevent the accumulation of toxic intermediates. Information exploited from such mutation-bred production strain would be useful for metabolic engineering.






Similar content being viewed by others
References
Archer JAC, Solow-Cordero DE, Sinskey AJ (1991) A C-terminal deletion in Corynebacterium glutumicum homoserine dehydrogenase abolishes allosteric inhibition by l-threonine. Gene 107:53–59
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C (2011) From zero to hero—Design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168
Bertani G (1951) Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300
Black S, Wright NG (1954) β-aspartate kinase and β-aspartyl-phosphate. J Biol Chem 213:27–38
Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ (2008) Corynebacterium glutamicum tailored for high-yield l-valine production. Appl Microbiol Biotechnol 79:471–479
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chassagnole C, Raїs B, Quentin E, Fell DA, Mazat J (2001) An integrated study of threonine-pathway enzyme kinetics in Escherichia coli. Biochem J 356:415–423
Chen Z, Meyer W, Rappert S, Sun J, Zeng AP (2011) Co-evolutionary analysis enabled rational deregulation of allosteric enzyme inhibition in Corynebacterium glutamicum for lysine production. Appl Environ Microbiol 77:4352–4360
Chen Z, Rappert S, Zeng AP (2015) Rational design of allosteric regulation of homoserine dehydrogenase by a nonnatural inhibitor l-lysine. ACS Synth Biol 4:126–131
Colón GE, Nguyen TT, Jetten MSM, Sinskey AJ, Stephanopoulos G (1995) Production of l-isoleucine by overexpression of ilvA in a Corynebacterium lactofermentum threonine producer. Appl Microbiol Biotechnol 43:482–488
de Jong BW, Shi S, Valle-Rodríguez JO, Siewers V, Nielsen J (2015) Metabolic pathway engineering for fatty acid ethyl ester production in Saccharomyces cerevisiae using stable chromosomal integration. J Ind Microbiol Biotechnol 42:477–486
Diesveld R, Tietze N, Fürst O, Reth A, Bathe B, Sahm H, Eggeling L (2009) Activity of exporters of Escherichia coli in Corynebacterium glutamicum, and their use to increase l-threonine production. J Mol Microbiol Biotechnol 16:198–207
Dong X, Quinn PJ, Wang X (2011) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for the production of l-threonine. Biotechnol Adv 29:11–23
Du J, Shao Z, Zhao H (2011) Engineering microbial factories for synthesis of value-added products. J Ind Microbiol Biotechnol 38:873–890
Eggeling I, Cordes C, Eggeling L, Sahm H (1987) Regulation of acetohydroxy acid synthase in Corynebacterium glutamicum during fermentation of α-ketobutyrate to l-isoleucine. Appl Microbiol Biotechnol 25:346–351
Eggeling L, Bott M (2005) Handbook of Corybacterium glutamicum. Taylor & Francis, Boca Raton
Eggeling L, Bott M (2015) A giant market and a powerful metabolism: l-lysine provided by Corybacterium glutamicum. Appl Microbiol Biotechnol 99:3387–3394
Follettie MT, Shin HK, Sinskey AJ (1988) Organization and regulation of the Corynebacterium glutamicum hom-thrB and thrC loci. Mol Microbiol 2:53–62
Guillouet S, Rodal AA, An G, Lessard PA, Sinskey AJ (1999) Expression of the Escherichia coli catabolic threonine dehydratase in Corynebacterium glutamicum and its effect on isoleucine production. Appl Environ Microbiol 65:3100–3107
Hasegawa S, Uematsu K, Natsuma Y, Suda M, Hiraga K, Jojima T, Inui M, Yukawa H (2012) Improvement of the redox balance increases l-valine production by Corynebacterium glutamicum under oxygen deprivation conditions. Appl Environ Microbiol 78(3):865–875
Hashiguchi K, Takesada H, Suzuki E, Matsui H (1999) Construction of an l-isoleucine overproducing strain of Escherichia coli K-12. Biosci Biotechnol Biochem 63:672–679
Holátko J, Elišáková V, Prouza M, Sobotka M, Nešvera J, Pátek M (2009) Metabolic engineering of the l-valine biosynthesis pathway in Corynebacterium glutamicum using promoter activity modulation. J Biotechnol 139:203–210
James CL, Viola RE (2002) Production and characterization of bifunctional enzymes. Domain swap** to produce new bifunctional enzymes in the aspartate pathway. Biochemistry 41:3720–3725
Koros A, Varga ZS, Molnar P (2008) Simultaneous analysis of amino acids and amines as their ophthalaldehyde-ethanethiol-9-fluorenylmethyl chloroformate derivatives in cheese by high-performance liquid chromatography. J Chromatogr A 1203:146–152
Liebl W, Bayerl A, Stillner U, Schleifer KH (1989) High efficiency electroporation of intact Corynebacterium glutamicum cells. FEMS Microbiol Lett 65:299–304
Miyajima R, Shiio I (1970) Regulation of aspartate family amino acid biosynthesis in Brevibacterium flavum. III. Properties of homoserine dehydrogenase. J Biochem 68:311–319
Möckel B, Eggeling L, Sahm H (1994) Threonine dehydratases of Corynebacterium glutamicum with altered allosteric control: their generation and biochemical and structural analysis. Mol Microbiol 13:833–842
Morbach S, Sahm H, Eggeling L (1995) Use of feedback-resistant threonine dehydratases of Corynebacterium glutamicum to increase carbon flux towards l-isoleucine. Appl Environ Microbiol 61:4315–4320
Morbach S, Sahm H, Eggeling L (1996) l-Isoleucine Production with Corynebacterium glutamicum: further flux increase and limitation of export. Appl Environ Microbiol 62:4345–4351
Park JH, Jang YS, Lee JW, Lee SY (2011) Escherichia coli W as a new platform strain for the enhanced production of l-valine by systems metabolic engineering. Biotechnol Bioeng 108:1140–1147
Park JH, Oh JE, Lee KH, Kim JY, Lee SY (2012) Rational design of Escherichia coli for l-isoleucine production. ACS Synth Biol 1:532–540
Park SD, Lee JY, Sim SY, Kim Y, Lee HS (2007) Characteristics of methionine production by an engineered Corynebacterium glutamicum strain. Metab Eng 9:327–336
Park JH, Lee KH, Kim TY, Lee SY (2007) Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico knockout simulation. Proc Natl Acad Sci USA 104:7797–7802
Park JH, Lee SY (2010) Fermentative production of branched chain amino acids: a focus on metabolic engineering. Appl Microbiol Biotechnol 85:491–506
Reinscheid DJ, Eikmanns BJ, Sahm H (1991) Analysis of a Corynebacterium glutamicum hom gene coding for a feedback-resistant homoserine dehydrogenase. J Bacteriol 173:3228–3230
Reinscheid DJ, Kronemeyer W, Eggeling L, Eikmanns BJ, Sahm H (1994) Stable expression of hom-1–thrB in Corynebacterium glutamicum and its effects on the carbon flux to threonine and related amino acids. Appl Environ Microbiol 60:126–132
Sahm H, Eggeling L, Morbach S (1999) Construction of l-isoleucine overproducing strains of Corynebacterium glutamicum. Naturwissenschaften 86:33–38
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Shiio I, Miyajima R (1969) Concerted inhibition and its reversal by end products of aspartate kinase in Brevibacterium flavum. J Biochem 64:849–859
Simic P, Willuhn J, Sahm H, Eggeling L (2002) Identification of glyA (encoding serine hydroxymethyl transferase) and its use together with the exporter ThrE to increase l-threonine accumulation by Corynebacterium glutamicum. Appl Environ Microbiol 68:3321–3327
Vogt M, Krumbach K, Bang WG, van Ooyen J, Noack S, Klein B, Bott M, Eggeling L (2015) The contest for precursors: channelling l-isoleucine synthesis in Corynebacterium glutamicum without byproduct formation. Appl Microbiol Biotechnol 99:791–800
Vogt M, Haas S, Klaffl S, Polen T, Eggeling L, van Ooyen J, Bott M (2014) Pushing product formation to its limit: metabolic engineering of Corynebacterium glutamicum for l-leucine overproduction. Metab Eng 22:40–52
Xu D, Tan Y, Huan X, Hu X, Wang X (2010) Construction of a novel shuttle vector for use in Brevibacterium flavum, an industrial amino acid producer. J Microbiol Methods 80:86–92
Wang J, Wen B, Wang J, Xu Q, Zhang C, Chen N, **e X (2013) Enhancing l-isoleucine production by thrABC overexpression combined with alaT deletion in Corynebacterium glutamicum. Appl Biochem Biotechnol 171:20–30
Yin L, Hu X, Xu D, Ning J, Chen J, Wang X (2012) Co-expression of feedback-resistant threonine dehydratase and acetohydroxy acid synthase increase l-isoleucine production in Corynebacterium glutamicum. Metab Eng 14:542–550
Yin L, Zhao J, Chen C, Hu X, Wang X (2014) Increasing l-isoleucine production in Corynebacterium glutamicum by enhancing the carbon flux in its biosynthesis pathway and NADPH supply. Biotechnol Bioprocess E 19:132–142
Yoshida A, Tomita T, Kurihara T, Fushinobu S, Kuzuyama T, Nishiyama M (2007) Structural insight into concerted inhibition of α2β2-type aspartate kinase from Corynebacterium glutamicum. J Mol Biol 368:521–536
Yoshida A, Tomita T, Kuzuyama T, Nishiyama M (2010) Mechanism of concerted inhibition of α2β2-type hetero-oligomeric aspartate kinase from Corynebacterium glutamicum. J Biol Chem 285:27477–27486
Yu X, ** H, Liu W, Wang Q, Qi Q (2015) Engineering Corynebacterium glutamicum to produce 5-aminolevulinic acid from glucose. Microb Cell Fact 14:183–192
Zhao J, Hu X, Li Y, Wang X (2015) Overexpression of ribosome elongation factor G and recycling factor increases l-isoleucine production in Corybacterium glutamicum. Appl Microbiol Biotechnol 99:4795–4805
Acknowledgments
This work was supported by National Natural Science Foundation of China (NSFC31370131), National Key Basic Research Program of China (973 Program 2012CB725202) and Fundamental Research Funds for the Central Universities (JUDCF11025). We thank Prof. Makoto Nishiyama (University of Tokyo) for providing information on inactivation of intrinsic ribosome binding site for the β subunit of AK, and Ms. Shuying Wang (Jiangnan University) for providing technical advice on HPLC analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dong, X., Zhao, Y., Zhao, J. et al. Characterization of aspartate kinase and homoserine dehydrogenase from Corynebacterium glutamicum IWJ001 and systematic investigation of l-isoleucine biosynthesis. J Ind Microbiol Biotechnol 43, 873–885 (2016). https://doi.org/10.1007/s10295-016-1763-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-016-1763-5