Abstract
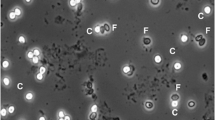
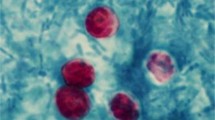
Cryptosporidiosis is an important though underreported public health concern. Molecular tools might be helpful in improving its diagnosis. In this study, ZR Fecal DNA MiniPrep™ Kit (ZR) and NucliSens® easyMAG® (EM) were compared using four Cryptosporidium-seeded feces and 29 Cryptosporidium-positive stools. Thereafter, ZR was selected for prospective evaluation of Cryptosporidium detection by 18S rDNA and LAXER quantitative PCR (qPCR) in 69 stools from 56 patients after Cryptosporidium detection by glycerin, modified Ziehl–Neelsen (ZN) and auramine–phenol (AP) stainings. The combination of any of the two extraction methods with 18S qPCR yielded adequate detection of Cryptosporidium in seeded stools, but the ZR kit showed the best performance. All 29 Cryptosporidium-positive samples were positive with 18S qPCR, after both ZR and EM extraction. However, false-negative results were found with LAXER qPCR or nested PCR. Cryptosporidiosis was diagnosed in 7/56 patients. All the microscopic methods enabled the initial diagnosis, but Cryptosporidium was detected in 12, 13, and 14 samples from these seven patients after glycerin, ZN, and AP staining respectively. Among these samples, 14 and 12 were positive with 18S and LAXER qPCR respectively. In two patients, Cryptosporidium DNA loads were found to be correlated with clinical evolution. Although little known, glycerin is a sensitive method for the initial detection of Cryptosporidium. When combined with 18S qPCR, ZR extraction, which had not been evaluated so far for Cryptosporidium, was an accurate tool for detecting Cryptosporidium and estimating the oocyst shedding in the course of infection.


Similar content being viewed by others
References
Ryan U, Paparini A, Tong K, Yang R, Gibson-Kueh S, O’Hara A et al (2015) Cryptosporidium huwi n. sp. (Apicomplexa: Eimeriidae) from the guppy (Poecilia reticulata). Exp Parasitol 150:31–35. doi:10.1016/j.exppara.2015.01.009
Ryan U, Fayer R, **ao L (2014) Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology 141:1667–1685. doi:10.1017/S0031182014001085
Šlapeta J (2013) Cryptosporidiosis and Cryptosporidium species in animals and humans: a thirty-colour rainbow? Int J Parasitol 43:957–970. doi:10.1016/j.ijpara.2013.07.005
Chalmers RM, Katzer F (2013) Looking for Cryptosporidium: the application of advances in detection and diagnosis. Trends Parasitol 29:237–251. doi:10.1016/j.pt.2013.03.001
Cacciò SM, Thompson RCA, McLauchlin J, Smith HV (2005) Unravelling Cryptosporidium and Giardia epidemiology. Trends Parasitol 21:430–437. doi:10.1016/j.pt.2005.06.013
Bouzid M, Hunter PR, Chalmers RM, Tyler KM (2013) Cryptosporidium pathogenicity and virulence. Clin Microbiol Rev 26:115–134. doi:10.1128/CMR.00076-12
Chalmers RM, Davies AP (2010) Minireview: clinical cryptosporidiosis. Exp Parasitol 124:138–146. doi:10.1016/j.exppara.2009.02.003
Hunter PR, Nichols G (2002) Epidemiology and clinical features of Cryptosporidium infection in immunocompromised patients. Clin Microbiol Rev 15:145–154
Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M et al (2015) A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94. doi:10.1016/S1473-3099(14)70772-8
Baldursson S, Karanis P (2011) Waterborne transmission of protozoan parasites: review of worldwide outbreaks — an update 2004–2010. Water Res 45:6603–6614. doi:10.1016/j.watres.2011.10.013
Robertson LJ, Chalmers RM (2013) Foodborne cryptosporidiosis: is there really more in Nordic countries? Trends Parasitol 29:3–9. doi:10.1016/j.pt.2012.10.003
Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S et al (2013) Burden and aetiology of diarrhoeal disease in infants and young children in develo** countries (the Global Enteric Multicenter Study, GEMS): a prospective, case–control study. Lancet 382:209–222. doi:10.1016/S0140-6736(13)60844-2
ANOFEL Cryptosporidium National Network (2010) Laboratory-based surveillance for Cryptosporidium in France, 2006–2009. Euro Surveill 15:19642
Henriksen SA, Pohlenz JF (1981) Staining of cryptosporidia by a modified Ziehl–Neelsen technique. Acta Vet Scand 22:594–596
Cacciò SM, Pozio E (2006) Advances in the epidemiology, diagnosis and treatment of cryptosporidiosis. Expert Rev Anti Infect Ther 4:429–443. doi:10.1586/14787210.4.3.429
Chalmers RM, Campbell BM, Crouch N, Charlett A, Davies AP (2011) Comparison of diagnostic sensitivity and specificity of seven Cryptosporidium assays used in the UK. J Med Microbiol 60:1598–1604. doi:10.1099/jmm.0.034181-0
Khurana S, Sharma P, Sharma A, Malla N (2012) Evaluation of Ziehl–Neelsen staining, auramine phenol staining, antigen detection enzyme linked immunosorbent assay and polymerase chain reaction, for the diagnosis of intestinal cryptosporidiosis. Trop Parasitol 2:20–23. doi:10.4103/2229-5070.97234
Jex AR, Smith HV, Monis PT, Campbell BE, Gasser RB (2008) Cryptosporidium—biotechnological advances in the detection, diagnosis and analysis of genetic variation. Biotechnol Adv 26:304–317. doi:10.1016/j.biotechadv.2008.02.003
Agnamey P, Sarfati C, Pinel C, Rabodoniriina M, Kapel N, Dutoit E et al (2011) Evaluation of four commercial rapid immunochromatographic assays for detection of Cryptosporidium antigens in stool samples: a blind multicenter trial. J Clin Microbiol 49:1605–1607. doi:10.1128/JCM.02074-10
Martín-Ampudia M, Mariscal A, Lopez-Gigosos RM, Mora L, Fernandez-Crehuet J (2012) Under-notification of cryptosporidiosis by routine clinical and laboratory practices among non-hospitalised children with acute diarrhoea in Southern Spain. Infection 40:113–119. doi:10.1007/s15010-011-0188-3
Calderaro A, Montecchini S, Gorrini C, Dettori G, Chezzi C (2011) Similar diagnostic performances of antigen detection and nucleic acid detection of Cryptosporidium spp. in a low-prevalence setting. Diagn Microbiol Infect Dis 70:72–77. doi:10.1016/j.diagmicrobio.2010.11.017
van Lieshout L, Roestenberg M (2015) Clinical consequences of new diagnostic tools for intestinal parasites. Clin Microbiol Infect 21:520–528. doi:10.1016/j.cmi.2015.03.015
**ao L, Escalante L, Yang C, Sulaiman I, Escalante AA, Montali RJ et al (1999) Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol 65:1578–1583
Hawash Y, Dorgham LS, Al-Hazmi AS, Al-Ghamdi MS (2014) Prevalence of Cryptosporidium-associated diarrhea in a high-altitude community of Saudi Arabia detected by conventional and molecular methods. Korean J Parasitol 52:479–485. doi:10.3347/kjp.2014.52.5.479
Abe N, Matsubayashi M, Kimata I, Iseki M (2006) Subgenotype analysis of Cryptosporidium parvum isolates from humans and animals in Japan using the 60-kDa glycoprotein gene sequences. Parasitol Res 99:303–305. doi:10.1007/s00436-006-0140-0
Homem CG, Nakamura AA, Silva DC, Teixeira WFP, Coelho WMD, Meireles MV (2012) Real-time PCR assay targeting the actin gene for the detection of Cryptosporidium parvum in calf fecal samples. Parasitol Res 110:1741–1745. doi:10.1007/s00436-011-2694-8
Laxer MA, Timblin BK, Patel RJ (1991) DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am J Trop Med Hyg 45:688–694
Tanriverdi S, Tanyeli A, Başlamişli F, Köksal F, Kilinç Y, Feng X et al (2002) Detection and genoty** of oocysts of Cryptosporidium parvum by real-time PCR and melting curve analysis. J Clin Microbiol 40:3237–3244
Sulaiman IM, Morgan UM, Thompson RC, Lal AA, **ao L (2000) Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl Environ Microbiol 66:2385–2391
Hawash Y (2014) DNA extraction from protozoan oocysts/cysts in feces for diagnostic PCR. Korean J Parasitol 52:263–271. doi:10.3347/kjp.2014.52.3.263
Halstead FD, Lee AV, Couto-Parada X, Polley SD, Ling C, Jenkins C et al (2013) Universal extraction method for gastrointestinal pathogens. J Med Microbiol 62:1535–1539. doi:10.1099/jmm.0.058743-0
Mary C, Chapey E, Dutoit E, Guyot K, Hasseine L, Jeddi F et al (2013) Multicentric evaluation of a new real-time PCR assay for quantification of Cryptosporidium spp. and identification of Cryptosporidium parvum and Cryptosporidium hominis. J Clin Microbiol 51:2556–2563. doi:10.1128/JCM.03458-12
Elwin K, Robinson G, Hadfield SJ, Fairclough HV, Iturriza-Gómara M, Chalmers RM (2012) A comparison of two approaches to extracting Cryptosporidium DNA from human stools as measured by a real-time PCR assay. J Microbiol Methods 89:38–40. doi:10.1016/j.mimet.2012.02.006
Masny A, Rozej W, Gołab E (2009) Development of efficient DNA isolation procedures for Cryptosporidium and Trichinella PCR detection in fecal samples. Med Dosw Mikrobiol 61:259–265
Dutoit E, Dewitte J-M, Dei-Cas E, Camus D (1988) Rapid method for detection of Cryptosporidium oocysts in stools. Le Biologiste XXII:159–160
Fontaine M, Guillot E (2002) Development of a TaqMan quantitative PCR assay specific for Cryptosporidium parvum. FEMS Microbiol Lett 214:13–17
Le Blancq SM, Khramtsov NV, Zamani F, Upton SJ, Wu TW (1997) Ribosomal RNA gene organization in Cryptosporidium parvum. Mol Biochem Parasitol 90:463–478
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. doi:10.1016/S0022-2836(05)80360-2
Potters I, Van Esbroeck M (2010) Negative staining technique of Heine for the detection of Cryptosporidium spp.: a fast and simple screening technique. Open Parasitol J 4:1–4
Kaushik K, Khurana S, Wanchu A, Malla N (2008) Evaluation of staining techniques, antigen detection and nested PCR for the diagnosis of cryptosporidiosis in HIV seropositive and seronegative patients. Acta Trop 107:1–7. doi:10.1016/j.actatropica.2008.02.007
Yoshikawa H, Dogruman-Al F, Dogruman-Ai F, Turk S, Kustimur S, Balaban N et al (2011) Evaluation of DNA extraction kits for molecular diagnosis of human Blastocystis subtypes from fecal samples. Parasitol Res 109:1045–1050. doi:10.1007/s00436-011-2342-3
Jothikumar N, da Silva AJ, Moura I, Qvarnstrom Y, Hill VR (2008) Detection and differentiation of Cryptosporidium hominis and Cryptosporidium parvum by dual TaqMan assays. J Med Microbiol 57:1099–1105. doi:10.1099/jmm.0.2008/001461-0
Fournet N, Deege MP, Urbanus AT, Nichols G, Rosner BM, Chalmers RM et al (2012) Simultaneous increase of Cryptosporidium infections in the Netherlands, the United Kingdom and Germany in late summer season, 2012. Euro Surveill 18:1–5
European Center for Disease Prevention and Control (ECDC) (2014) Annual epidemiological report 2014. Food- and waterborne diseases and zoonoses
Menotti J, Cassinat B, Porcher R, Sarfati C, Derouin F, Molina J-M (2003) Development of a real-time polymerase-chain-reaction assay for quantitative detection of Enterocytozoon bieneusi DNA in stool specimens from immunocompromised patients with intestinal microsporidiosis. J Infect Dis 187:1469–1474. doi:10.1086/374620
Mouthon L, Lortholary O (2003) Intravenous immunoglobulins in infectious diseases: where do we stand? Clin Microbiol Infect 9:333–338. doi:10.1046/j.1469-0691.2003.00694.x
Acknowledgments
We wish to thank Michèle Wauquier and Filoména Naji (Parasitology–Mycology Laboratory of Lille University Hospital Center) for their technical assistance, and the members of the French ANOFEL Cryptosporidium National Network: Ahmed Abou Bacar CHU Strasbourg, Isabelle Accoceberry CHU Bordeaux, Patrice Agnamey CHU Amiens, Adela Angoulvant CHU Bicêtre Paris, Dominique Aubert CHU Amiens, Belkhadi Ghania CHU St Antoine, Paris, Antoine Berry CHU Toulouse, Denis Blanchet CHU Cayenne, Julie Bonhomme CHU Caen, Françoise Botterel CHU Créteil, Marie-Elisabeth Bougnoux CHU Necker, Paris, Pierre Buffet CHU Pitié, Paris, Frédéric Dalle CHU Dijon, Eric Dannaoui HEGP, Paris, Marie-Laure Dardé CHU Limoges, Ludovic De Gentile CHU Angers, Anne Debourgogne CHU Nancy, Monique Debruyne (Cerba, Paris), Brigitte Degeilh CHU Rennes, Magalie Demar CHU Cayenne, Nicole Desbois CHU Fort de France, Guillaume Desoubeaux CHU Tours, Pascal Delaunay CHU Nice, Pierre Flori CHU St Etienne, Gilles Gargala CHU Rouen, Agathe Goubard Biomnis Paris, Frédéric Grenouillet CHU Besançon, Samia Hamane CHU St Louis, Paris, Sandrine Houzé CHU Bichat, Paris, Jamet Deborah CHU Brest, Nathalie Kapel CHU Pitié, Paris, Franck Labbe CH Le Havre, Denis Leméteil Lab. St Valéry en Caux, Denis Magne CHU St Antoine Paris, Pierre Marty CHU Nice, Jean Menotti CHU St Louis, Paris, Laurence Millon CHU Besançon, Christelle Morelle CHU Montpellier, Florent Morio CHU Nantes, Jean-Benjamin Murat CHU Grenoble, Gilles Nevez CHU Brest, Muriel Nicolas CHU Guadeloupe, Philippe Poirier CHU Clermont-Ferrand, Méja Rabodonirina CHU Lyon, Marie-Hélène Rodier CHU Poitiers, Marc Sautour CHU Dijon, Marc Thellier CHU Pitié, Paris, Anne Totet CHU Amiens, Alexis Valentin CHU Toulouse, Isabelle Villena CHU Reims, Hélène Yera CHU Cochin, Paris.
This work was supported by grants from the Lille University Hospital Center, from the University of Lille, Pasteur Institute of Lille, “Centre National de la Recherche Scientifique” (CNRS), and “Institut National de la Santé et de la Recherche Médicale” (Inserm). It was presented at the First French North African Parasitology and Mycology meeting in Rabat, Morocco, in October 2013, and at the European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) in Barcelona, Spain, in May 2014.
Author information
Authors and Affiliations
Consortia
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Eduardo Dei-Cas is deceased.
E. Fréalle and E. Dutoit contributed equally to this work.
Rights and permissions
About this article
Cite this article
Le Govic, Y., Guyot, K., Certad, G. et al. Assessment of microscopic and molecular tools for the diagnosis and follow-up of cryptosporidiosis in patients at risk. Eur J Clin Microbiol Infect Dis 35, 137–148 (2016). https://doi.org/10.1007/s10096-015-2519-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2519-2