Abstract
Context
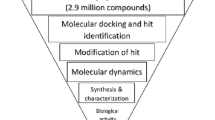
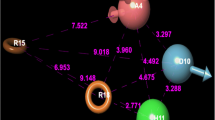
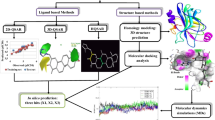
Protein–protein interaction interfaces play a major role in cell signaling pathways. There is always a great interest in develo** protein–protein interaction (PPI) inhibitors of kinases, as they are challenging due to their hydrophobicity, flat surface, specificity, potency, etc. 3 Phosphoinositide-dependent kinase-1 (PDK1), which is involved in the PI3K/PDK1/AKT pathway, is a cancer target that has gained insight for the past two decades. PDK1 possesses a protein interaction fragment (PIF) pocket, which is a well-known PPI that targets allosteric modulators. This work focusses on energy-based pharmacophore model development which on virtual screening could yield novel scaffolds towards the drug designing objective for the kind of PDK1 modulators. A novel pyrazolo pyridine molecule was identified as an allosteric modulator that binds to the PPI site. The metadynamics simulations with free energy profiles further revealed the conformational allosteric changes stimulated on the protein structure upon ligand binding. The cytotoxic activity (IC50-20 μM) of the identified compound against five different cancer cell lines and cell cycle analysis supported the anticancer activity of the identified compound.
Methods
All the computational works were carried out by the most commonly used Schrodinger Suite software. The pharmacophore was validated by Receiver Operation Characteristics (ROC) and screening against allosteric Enamine database library. The Optimized Potential Liquid Simulations (OPLS-2005) was used to minimize the structures for molecular docking calculations, and inbuilt scoring method of ranking the compounds based on docking score and Glide energy was used. Molecular dynamics simulations were conducted by Desmond implemented in Maestro. The hit compound was purchased from Enamine and tested against different cancer cell lines by MTT assay, apoptosis by western blotting technique, and by flow cytometry analysis.









Similar content being viewed by others
References
Shekawat SS, Ghosh I (2011) Split-protein systems: beyond binary protein-protein interactions. Curr Opin Chem Biol 15(6):789–797. https://doi.org/10.17159/tvl.v.53i2.27
Bakail M, Ochsenbein F (2016) Targeting protein-protein interactions, a wide-open field for drug design. C R Chim 19(1–2):19–27. https://doi.org/10.1016/j.crci.2015.12.004
Cossar PJ, Lewis PJ, McCluskey A (2020) Protein-protein interactions as antibiotic targets: a medicinal chemistry perspective. Med Res Rev 40(2):469–494. https://doi.org/10.1002/med.21519
** L, Wang W, Fang G (2014) Targeting protein-protein interactions by small molecules. Annu Rev Pharmacol Toxicol 54:435–456. https://doi.org/10.1007/978-981-13-0773-7
Wang X, Ni D, Liu Y, Lu S (2021) Rational design of peptide-based inhibitors disrupting protein-protein interactions. Front Chem 9(May):1–15. https://doi.org/10.3389/fchem.2021.682675
Marondedze EF, Govender KK, Govender PP (2020) Ligand-based pharmacophore modelling and virtual screening for the identification of amyloid-beta diagnostic molecules. J Mol Graph Model 101:107711. https://doi.org/10.1016/j.jmgm.2020.107711
Abell C, Scott DE, Ehebauer MT, Pukala T, Marsh M, Blundell TL, Venkitaraman AR, Hyvönen M (2013) Using a fragment-based approach to target protein-protein interactions. ChemBioChem 14(3):332–342. https://doi.org/10.1002/cbic.201200521
Petta I, Lievens S, Libert C, Tavernier J, De Bosscher K (2016) Modulation of protein-protein interactions for the development of novel therapeutics. Mol Ther 24(4):707–718. https://doi.org/10.1038/mt.2015.214
Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, Huang DCS, Hymowitz SG, ** S, Khaw SL, Kovar PJ, Lam LT, Lee J, Maecker HL, Marsh KC et al (2013) ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19(2):202–208. https://doi.org/10.1038/nm.3048
Gagliardi PA, Puliafito A, Primo L (2018) PDK1: at the crossroad of cancer signaling pathways. Semin Cancer Biol 48:27–35. https://doi.org/10.1016/j.semcancer.2017.04.014
Harris TK (2003) PDK1 and PKB/Akt: ideal targets for development of new strategies to structure-based drug design. IUBMB Life 55(3):117–126. https://doi.org/10.1080/1521654031000115951
Najafov A, Sommer EM, Axten JM, Deyoung MP, Alessi DR (2011) Characterization of GSK2334470, a novel and highly specific inhibitor of PDK1. Biochem J 433(2):357–369. https://doi.org/10.1042/BJ20101732
Biondi RM, Komander D, Thomas CC, Lizcano JM, Deak M, Alessi DR, van Aalten DMF (2002) High resolution crystal structure of the human PDK1 catalytic domain defines the regulatory phosphopeptide docking site. EMBO J 21(16):4219–4228. https://doi.org/10.1093/emboj/cdf437
Xu X, Chen Y, Fu Q, Ni D, Zhang J, Li X, Lu S (2019) The chemical diversity and structure-based discovery of allosteric modulators for the PIF-pocket of protein kinase PDK1. J Enzyme Inhib Med Chem 34(1):361–374. https://doi.org/10.1080/14756366.2018.1553167
Arkin MMR, Wells JA (2004) Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov 3(4):301–317. https://doi.org/10.1038/NRD1343
Rettenmaier TJ, Sadowsky JD, Thomsen ND, Chen SC, Doak AK, Arkin MR, Wells JA (2014) A small-molecule mimic of a peptide docking motif inhibits the protein kinase PDK1. Proc Natl Acad Sci 111(52):18590–18595. https://doi.org/10.1073/pnas.1415365112
Arkin MR, Tang Y, Wells JA (2014) Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21(9):1102–1114. https://doi.org/10.1016/j.chembiol.2014.09.001
Engel M, Hindie V, Lopez-Garcia LA, Stroba A, Schaeffer F, Adrian I, Imig J, Idrissova L, Nastainczyk W, Zeuzem S, Alzari PM, Hartmann RW, Piiper A, Biondi RM (2006) Allosteric activation of the protein kinase PDK1 with low molecular weight compounds. EMBO J 25(23):5469–5480. https://doi.org/10.1038/sj.emboj.7601416
Schulze JO, Saladino G, Busschots K, Neimanis S, Süß E, Odadzic D, Zeuzem S, Hindie V, Herbrand AK, Lisa MN, Alzari PM, Gervasio FL, Biondi RM (2016) Bidirectional allosteric communication between the ATP-binding site and the regulatory PIF pocket in PDK1 protein kinase. Cell Chemical Biology 23(10):1193–1205. https://doi.org/10.1016/j.chembiol.2016.06.017
Sestito S, Rapposelli S (2019) A patent update on PDK1 inhibitors (2015-present). Expert Opin Ther Pat 29(4):271–282. https://doi.org/10.1080/13543776.2019.1597852
Sadowsky JD, Burlingame MA, Wolan DW, McClendon CL, Jacobson MP, Wells JA (2011) Turning a protein kinase on or off from a single allosteric site via disulfide trap**. Proc Natl Acad Sci U S A 108(15):6056–6061. https://doi.org/10.1073/pnas.1102376108
Beekman AM, Cominetti MMD, Walpole SJ, Prabhu S, O’Connell MA, Angulo J, Searcey M (2019) Identification of selective protein-protein interaction inhibitors using efficient: in silico peptide-directed ligand design. Chem Sci 10(16):4502–4508. https://doi.org/10.1039/c9sc00059c
Gonzalez-Ruiz D, Gohlke H (2006) Targeting protein-protein interactions with small molecules: challenges and perspectives for computational binding epitope detection and ligand finding. Curr Med Chem 13(22):2607–2625. https://doi.org/10.2174/092986706778201530
Li B, Rong D, Wang Y (2019) Targeting protein-protein interaction with covalent small-molecule inhibitors. Curr Top Med Chem 19(21):1872–1876
Sable R, Jois S (2015) Surfing the protein-protein interaction surface using docking methods: application to the design of PPI inhibitors. Molecules 20(6):11569–11603. https://doi.org/10.3390/molecules200611569
Sarvagalla S, Cheung CHA, Tsai JY, Hsieh HP, Coumar MS (2016) Disruption of protein-protein interactions: hot spot detection, structure-based virtual screening and: In vitro testing for the anti-cancer drug target-survivin. RSC Adv 6(38):31947–31959. https://doi.org/10.1039/c5ra22927h
Cohen P, Cross D, Jänne PA (2021) Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov 20(7):551–569. https://doi.org/10.1038/s41573-021-00195-4
Dixon SL, Smondyrev AM, Rao SN (2006) PHASE: a novel approach to pharmacophore modeling and 3D database searching. Chem Biol Drug Des 67(5):370–372. https://doi.org/10.1111/j.1747-0285.2006.00384.x
Schrödinger Release 2017-1: Phase, Schrödinger, LLC, New York, NY, 2017
Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY (2021) Maestro-Desmond interoperability tools. Schrödinger, New York, NY, p 2021
Li G, Wang Z, Chong T, Yang J, Li H, Chen H (2017) Curcumin enhances the radiosensitivity of renal cancer cells by suppressing NF-κB signaling pathway. Biomed Pharmacother 94:974–981. https://doi.org/10.1016/J.BIOPHA.2017.07.148
Rettenmaier TJ, Fan H, Karpiak J, Doak A, Sali A, Shoichet BK, Wells JA (2015) Small-molecule allosteric modulators of the protein kinase PDK1 from structure-based docking. J Med Chem 58(20):8285–8291. https://doi.org/10.1021/acs.jmedchem.5b01216
Liu W, Li P, Mei Y (2019) Discovery of SBF1 as an allosteric inhibitor targeting the PIF-pocket of 3-phosphoinositide-dependent protein kinase-1. J Mol Model 25(7). https://doi.org/10.1007/s00894-019-4069-5
Wang J, Peng C, Yu Y, Chen Z, Xu Z, Cai T et al (2020). Biophys J 118:1009
Stroba A, Schaeffer F, Hindie V, Lopez-Garcia L, Adrian I, Fröhner W et al (2009). J Med Chem 52:4683
Busschots K, Lopez-Garcia LA, Lammi C, Stroba A, Zeuzem S, Piiper A et al (2012). Chem Biol 19:1152
Stakheev D, Taborska P, Strizova Z, Podrazil M, Bartunkova J, Smrz D (2019). Sci Rep 9(1):4761
Altamimi AS, El-Azab AS, Abdelhamid SG, Alamri MA, Bayoumi AH, Alqahtani SM, Alabbas AB, Altharawi AI, Alossaimi MA, Mohamed MA (2021). Molecules 26(10):2992
Chen S, Ye H, Gong F, Mao S, Li C, Xu B, Ren Y, Yu R (2021). Oncol Rep 45(4):38
Acknowledgements
KNV greatly acknowledges the Council for Scientific and Industrial Research (CSIR), Govt. of India for a Senior Research Associate fellowship through the Scientists’ Pool scheme. The authors also acknowledge Prof. Sung-Hoon Kim’s laboratory, College of Korean Medicine, Kyunghee University, South Korea, for all the in vitro studies.
Author information
Authors and Affiliations
Contributions
Both authors equally contributed to the work and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kailasam Natesan, V., Kuppannagounder Pitchaimuthu, E. Structure-based drug design and molecular dynamics studies of an allosteric modulator targeting the protein–protein interaction site of PDK1. J Mol Model 30, 51 (2024). https://doi.org/10.1007/s00894-024-05842-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-024-05842-2