Abstract
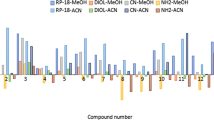
Retention behaviour of twenty-one chalcones synthesized in our laboratory was tested in three thin-layer chromatography (RP-TLC) systems (acetonitrile–water, ethanol–water and acetone–water) and chromatography parameters \( {R}_M^0 \), S and C0 were calculated. The most suitable RP-TLC system (acetonitrile–water) and chromatography parameter (C0) for lipophilicity prediction of tested compounds were selected on the basis of the highest correlations with calculated logP values. In selected system, compound 12 had the highest, whereas 47 had the lowest C0 value. QSRR analysis was performed and three models representing relationships between C0 and selected molecular descriptors were created—MLR(C0), PLS(C0) and SVM(C0). Interpretation of molecular descriptors which form statistically the most reliable SVM(C0) model identified the most important structural and physico-chemical properties that influence retention behaviour of tested compounds. In addition, descriptors with the highest influence on \( {R}_M^0 \) as well as on C0 calculated in the remaining two RP-TLC systems were identified and interpreted.




Similar content being viewed by others
References
Morak-Młodawska E, Nowak M, Pluta K (2007) Determination of the lipophilicity parameters RM0 and LogP of new azaphenothiazines by reversed-phase thin-layer chromatography. J Liq Chromatogr R T 30:1845–1854. https://doi.org/10.1080/10826070701360749
Csermely T, Kalász H, Deák K, Mohammed Y, Hasan MY, Darvas F, Petroianu G (2008) Lipophilicity determination of some ACE inhibitors by TLC. J Liq Chromatogr R T 31:2019–2034. https://doi.org/10.1080/10826070802198410
Odović J, Karljiković-Rajić K, Trbojević-Stanković J, Stojimirović B, Vladimirov S (2012) Lipophilicity examination of some ACE inhibitors and hydrochlorothiazide on cellulose in RP thin-layer chromatography. Iran J Pharm Res 11:763–770
Dobričić V, Vladimirov S, Čudina O (2016) Synthesis and RP-TLC lipophilicity evaluation of a novel fluocinolon acetonide soft drug derivative. Kragujevac J Sci 38:107–114. https://doi.org/10.5937/KgJSci1638107D
**a Y, Yang Z-Y, **a P, Bastow KF, Nakanishi Y, Lee K-H (2000) Antitumor agents. Part 202: novel 2′-amino chalcones: design, synthesis and biological evaluation. Bioorg Med Chem Lett 10:699–701. https://doi.org/10.1016/s0960-894x(00)00072-x
Venkatesan P, Sumathi S (2010) Piperidine mediated synthesis of n-heterocyclic chalcones and their antibacterial activity. J Heterocyclic Chem 47:81–84. https://doi.org/10.1002/jhet.268
Gomes MN, Muratov EN, Pereira M, Peixoto JC, Rosseto LP, Cravo PVL, Andrade CH, Neves BJ (2017) Chalcone derivatives: promising starting points for drug design. Molecules 22:E1210. https://doi.org/10.3390/molecules22081210
Turkovic N, Ivkovic B, Kotur-Stevuljevic J, Tasic M, Marković B, Vujic Z (2020) The molecular docking, synthesis and anti-HIV-1 protease activity of novel chalcones. Curr Pharm Des 26:802–814. https://doi.org/10.2174/1381612826666200203125557
Soczewiński E, Wachtmeister CA (1962) The relation between the composition of certain ternary two-phase solvent systems and RM values. J Chromatogr A 7:311–320. https://doi.org/10.1016/S0021-9673(01)86422-0
CambridgeSoft Corporation. 2005. ChemDraw ultra version 8.0.3. Cambridge, MA, USA
MarvinSketch 15.1.26 (2015). ChemAxon, Budapest. http://www.chemaxon.com
Ghose AK, Crippen GM (1987) Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2. Modeling dispersive and hydrophobic interactions. J Chem Inf Comput Sci 27:21–35. https://doi.org/10.1021/ci00053a005
Viswanadhan VN, Ghose AK, Revankar GR, Robins RK (1989) Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J Chem Inf Comput Sci 29:163–172. https://doi.org/10.1021/ci00063a006
Broto P, Moreau G, Vandycke C (1984) Molecular structures: perception, autocorrelation descriptor and SAR studies. System of atomic contributions for the calculation of n-octane/water partition coefficients. Eur J Med Chem- Chim Theor 19:71–78
Dong J, Cao DS, Miao HY, Liu S, Deng B-C, Yun Y-H, Wang N-N, Lu AP, Zeng W-B, Chen AF (2015) ChemDes: an integrated web-based platform for molecular descriptor and fingerprint computation. Aust J Chem 7:60. https://doi.org/10.1186/s13321-015-0109-z
TIBCO Software Inc., 2017. Statistica (data analysis software system), version 13. http://statistica.io
Gupta VK, Khani H, Ahmadi-Roudi B, Mirakhorli S, Fereyduni E, Agarwal S (2011) Prediction of capillary gas chromatographic retention times of fatty acid methyl esters in human blood using MLR, PLS and back-propagation artificial neural networks. Talanta 83:1014–1022. https://doi.org/10.1016/j.talanta.2010.11.017
Zhang YX (2007) Artificial neural networks based on principal component analysis input selection for clinical pattern recognition analysis. Talanta 73:68–75. https://doi.org/10.1016/j.talanta.2007.02.030
Gonzalez-Arjona D, Lopez-Perez G, Gustavo-Gonzalez A (2002) Non-linear QSAR modeling by using multilayer perceptron feedforward neural networks trained by back-propagation. Talanta 56:79–90. https://doi.org/10.1016/S0039-9140(01)00537-9
Dobričić V, Marković B, Nikolic K, Savić V, Vladimirov S, Čudina O (2014) 17β-carboxamide steroids--in vitro prediction of human skin permeability and retention using PAMPA technique. Eur J Pharm Sci 52:95–108. https://doi.org/10.1016/j.ejps.2013.10.017
Cortes C, Vapnik V (1995) Support-vector networks. Mach Learn 20:273–297. https://doi.org/10.1007/BF00994018
Heitkamp K, Bajorath J (2014) Support vector machines for drug discovery. Expert Opin Drug Discovery 9:93–104. https://doi.org/10.1517/17460441.2014.866943
Vucicevic J, Popovic M, Nikolic K, Filipic S, Obradovic D, Agbaba D (2017) Use of biopartitioning micellar chromatography and RP-HPLC for the determination of blood–brain barrier penetration of α-adrenergic/imidazoline receptor ligands, and QSPR analysis. SAR QSAR Environ Res 28:235–252. https://doi.org/10.1080/1062936X.2017.1302506
Ojha PK, Roy K (2011) Comparative QSARs for antimalarial endochins: importance of descriptor-thinning and noise reduction prior to feature selection. Chemometr Intell Lab 109:146–161. https://doi.org/10.1016/j.chemolab.2011.08.007
Snedecor GW, Cochran WG (1967) Statistical methods. Oxford & IBH, New Delhi
Marshall GR (1994) Binding-site modeling of unknown receptors. In: Kubinyi H (ed) 3D QSAR in drug design – theory, methods and applications. ESCOM, Leiden, pp 80–116
Tropsha A (2010) Best practices for QSAR model development, validation and exploitation. Mol Inf 29:476–488. https://doi.org/10.1002/minf.201000061
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstrom C, Wold S (2001) Multi-and Megavariate Data Analysis Basic Principles And Applications I, 2nd edn. Umetrics Academy, Umeå
Kuchar M, Kraus E, Jelínková M (1991) Influence of mobile phase composition on evaluation of lipophilicity by partition chromatography. J Chromatogr 557:399–411
Starek M, Komsta Ł, Krzek J (2013) Reversed-phase thin-layer chromatography technique for the comparison of the lipophilicity of selected non-steroidal anti-inflammatory drugs. J Pharm Biomed Anal 85:132–137. https://doi.org/10.1016/j.jpba.2013.07.017
Tesic ZL, Milojkovic-Opsenica DM (2013) TLC determination of drug lipophilicity. In: Komsta L, Waksmundzka-Hajnos M, Sherma J (eds) Thin Layer Chromatography in Drug Analysis. CRC Press, Taylor & Francis Group, Boca Raton, pp 225–246
Biagi GL, Barbaro AM, Sapone A, Recanatini M (1994) Determination of lipophilicity by means of reversed-phase thin-layer chromatography: I. Basic aspects and relationship between slope and intercept of TLC equations. J Chromatogr A 662:341–361. https://doi.org/10.1016/0021-9673(94)80521-0
Rageh AH, Atia NN, Abdel-Rahman HM (2017) Lipophilicity estimation of statins as a decisive physicochemical parameter for their hepato-selectivity using reversed-phase thin layer chromatography. J Pharm Biomed Anal 142:7–14. https://doi.org/10.1016/j.jpba.2017.04.037
Morak B, Nowak M, Pluta K (2007) Determination of the lipophilicity parameters R M0 and log P of new azaphenothiazines by reversed-phase thin-layer chromatography. J Liq Chromatogr R T 30:1845–1854
Ciura K, Fedorowicz J, Andrić F, Greber KE, Gurgielewicz A, Sawicki W, Sączewski J (2019) Lipophilicity determination of quaternary (fluoro) quinolones by chromatographic and theoretical approaches. Int J Mol Sci 20:E5288. https://doi.org/10.3390/ijms20215288
Constantinescu T, Lungu CN, Lung I (2019) Lipophilicity as a central component of drug-like properties of chalchones and flavonoid derivatives. Molecules 24:E1505. https://doi.org/10.3390/molecules24081505
Ciura K, Nowakowska J, Pikul P, Struck-Lewicka W, Markuszewski MJ (2015) A comparative quantitative structure-retention relationships study for lipophilicity determination of compounds with a phenanthrene skeleton on cyano-, reversed phase-, and normal phase-thin layer chromatography stationary phases. J AOAC Int 98:345–353. https://doi.org/10.5740/jaoacint.14-187
Burden FR (1989) Molecular identification number for substructure searches. J Chem Inf Comput Sci 29:225–227. https://doi.org/10.1021/ci00063a011
Burden FR (1997) A chemically intuitive molecular index based on the eigenvalues of a modified adjacency matrix. Quant Struct-Act Relat 16:309–314. https://doi.org/10.1002/qsar.19970160406
Kang YK, Jhon MS (1982) Additivity of atomic static polarizabilities and dispersion coefficients. Theor Chim Acta 61:41–48. https://doi.org/10.1007/BF00573863
Takaku T, Nagahori H, Sogame Y, Takagi T (2015) Quantitative structure–activity relationship model for the fetal–maternal blood concentration ratio of chemicals in humans. Biol Pharm Bull 38:930–934. https://doi.org/10.1248/bpb.b14-00883
Hajimahdi Z, Safizadeh F, Zarghi A (2016) QSAR analysis for some 1, 2-benzisothiazol-3-one derivatives as caspase-3 inhibitors by stepwise MLR method. Iran J Pharm Res IJPR 15:439–448
Madden JC (2011) Introduction to QSAR and other in silico methods to predict toxicity. In: Cronin MTD, Madden JC (eds) In silico toxicology: principles and applications, RSC Publishing, pp 11-30
Availability of data and material
All raw data are available upon request.
Funding
This work was financially supported by the Ministry of Education, Science and Technological Development, Republic of Serbia, as part of Project No.172041.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Dobričić, V., Turković, N., Ivković, B. et al. Evaluation of the lipophilicity of chalcones by RP-TLC and computational methods. JPC-J Planar Chromat 33, 245–253 (2020). https://doi.org/10.1007/s00764-020-00029-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00764-020-00029-w