Abstract
Orpheoviruses, cedratviruses, and pithoviruses are large DNA viruses that cluster together taxonomically within the order Pimascovirales of the phylum Nucleocytoviricota. However, they were not classified previously by the International Committee on Taxonomy of Viruses (ICTV). Here, we present a comprehensive analysis of the gene content, morphology, and phylogenomics of these viruses, providing data that underpinned the recent proposal to establish new taxa for their initial classification. The new taxonomy, which has now been ratified by the ICTV, includes the family Orpheoviridae and genus Alphaorpheovirus, the family Pithoviridae and genus Alphapithovirus, and the family Cedratviridae and genus Alphacedratvirus, aiming to formally catalogue the isolates covered in this study. Additionally, as per the newly adopted rules, we applied standardized binomial names for the virus species created to classify isolates with complete genome sequences available in public databases at the time of the proposal. The specific epithet of each virus species was chosen as a reference to the location where the exemplar virus was isolated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the first decade after the discovery of amoeba-infecting giant viruses in 2003, two taxonomic families were created due to their structural and genomic characteristics, which differed significantly from those of other viruses known then. These families, Mimiviridae and Marseilleviridae, were created to classify representatives of the only two taxa that had been established at the time. However, with the increased interest and intensification of research on this topic in the last 10 years, several other groups of giant amoeba viruses have been discovered [1,2,3,4,5,6,7,8,9,10]. Despite recent progress in taxonomy of giant viruses, many of them have not yet been formally assigned to any taxon recognized by the ICTV [11,12,13,14].
Despite of their great diversity, these viruses are hypothetically monophyletic and are grouped within the phylum Nucleocytoviricota. Within this phylum, the giant amoeba viruses recognized to date have been classified in the class Megaviricetes. Within this class, the order Pimascovirales previously included three recognized families: Ascoviridae and Iridoviridae, which have invertebrates and some groups of vertebrates as hosts, and Marseilleviridae, which comprises amoeba-infecting giant viruses [15]. However, three related groups of ovoid amoeba-infecting giant viruses had not yet received formal taxonomic recognition. These groups are the pithoviruses, cedratviruses, and orpheoviruses.
Despite having similar shapes and sizes, these three groups of viruses have many morphological and genomic differences that strongly distinguish them from other families within this order and from each other [16]. Therefore, in order to start the process of official classification of these isolates within the ICTV taxonomy framework, we proposed the formal creation of seven species: two for the pithoviruses, four for the cedratviruses, and one for the orpheoviruses, to be classified in the newly established genera “Alphapithovirus”, “Alphacedratvirus”, and “Alphaorpheovirus”, belonging to the new families “Pithoviridae”, “Cedratviridae”, and “Orpheoviridae”, respectively.
Etymology of taxon nomenclature
The family Pithoviridae and its representatives
We formally proposed to create the family “Pithoviridae”, composed of a new genus, “Alphapithovirus”, and two new species, “Alphapithovirus sibericum” and “Alphapithovirus massiliense”. “Pitho-” is derived from the Greek pithos, a kind of large amphora. The names of the species refer to the names of the founding isolates already used in the literature, with “alpha” as a prefix to denote the genus in which the original isolate is classified. The epithets were derived from the informal names initially proposed by the original authors who discovered the viruses and subsequently modified to Latinized singular forms, as exemplified by the standardized classification of the amoebal virus species Yaravirus brasiliense [2, 13, 17, 18].
The family Cedratviridae and its representatives
We also proposed to create the family “Cedratviridae”, composed of a new genus, “Alphacedratvirus”, and four species within this genus: “Alphacedratvirus aljazairense”, comprising the isolate cedratvirus A11, “Alphacedratvirus franciense”, comprising cedratvirus lausannensis and cedratvirus zaza, “Alphacedratvirus brasiliense”, comprising Brazilian cedratvirus, and “Alphacedratvirus rossiense” comprising cedratvirus kamchatka. Once more, the particle morphology served as an inspiration for the naming: “Cedrat-” from French cédrat, citron [3, 19,20,21].
The first cedratvirus, named cedratvirus A11, was isolated in 2016 from water samples collected in different regions of Algeria. To standardize the nomenclature, we propose naming the species, founded by the isolate cedratvirus A11, “Alphacedratvirus aljazairense”. “Aljazair-” is the Romanized form of the name Algeria in Arabic, al-Jazāʾir. The other proposed species names also follow the same pattern, derived from the geographic locations where isolates were originally collected. In the following years, other cedratviruses were isolated from water samples from France (cedratvirus lausannensis and cedratvirus zaza) and Brazil (Brazilian cedratvirus) and a soil sample collected in Russia (cedratvirus kamchatka) [3, 19,20,21].
The family Orpheoviridae and its representative
Lastly, we proposed the creation of the species “Alphaorpheovirus massiliense”, the genus “Alphaorpheovirus”, and the family “Orpheoviridae” to classify one viral isolate named orpheovirus IHUMI-LCC2. “Orpheo-” was inspired by the Greek legend of Orpheus [1].
Morphological properties
Instead of exhibiting icosahedral capsid symmetry, pithoviruses, cedratviruses, and orpheovirus have unconventional virion morphologies that are similar to each other. They feature elongated, ovoid-shaped particles measuring approximately 1 µm in length, but these viruses can be distinguished morphologically by the arrangement and number of delivery portals present in their particles. Whilst cedratviruses contain two corks, one on each apex of the particle, pithoviruses more often contain only one cork, and orpheovirus has one ostiole, also located on the apex of the particle. The delivery portals can often be observed by transmission electron microscopy [1, 3, 17, 22,23,24].
Occurrence and host range
The representatives of this clade have been isolated on at least four different continents. Over 10 cedratvirus isolates and four pithovirus isolates have been discovered using Acanthamoeba sp. as a cellular support host, but so far, only one orpheovirus isolate has been discovered, using Vermamoeba vermiformis [1,2,3, 17, 19,20,21, 23, 25,26,27,28,29,30,31]. These viruses have been found in a diverse array of samples, obtained from rivers, rat feces, permafrost, sewage water, and muddy soil. Despite the increased prospecting efforts in recent years, metagenomics work indicates that this clade exhibits significant diversity, with many different hierarchical taxa within the order Pimascovirales that remain largely unexplored [32,33,34,35].
Comparative genomic features
Viruses of this clade exhibit a large amount of variation in the size of their genomes, GC content, and coding density, but when compared at the family level, the characteristics become fairly consistent among the isolates. The main characteristics of the circular dsDNA genomes of these viruses are listed in Table 1.
The same pattern can be observed in the synteny analysis. A whole-genome alignment made using Mauve software showed strong synteny between the pithoviruses. Similar genomic arrangements could also be observed within each lineage of cedratvirus, with cedratvirus A11, cedratvirus zaza, and cedratvirus lausannensis belonging to lineage A and cedratvirus kamchatka and Brazilian cedratvirus to lineage B. However, despite the clear synteny between pithoviruses and cedratviruses when orpheovirus was added to the analysis, major differences were observed in terms of genome size, composition, and genetic organization (Fig. 1).
Genomic synteny analysis. A comparison of the genome organization of members of the three new families “Cedratviridae”, “Orpheoviridae”, and “Pithoviridae” is shown. The schematic diagram was produced using the Mauve software package [36]
The pairwise average nucleotide identity (ANI) and average amino acid identity (AAI) for pithoviruses were > 84% for the two isolates, and their gene sharing level was 78%. The pairwise ANI and AAI values for all of the cedratviruses was > 70%, and their gene sharing level was > 67%. An intergroup comparison between the proposed families showed a pairwise ANI < 30%. The average pairwise AAI and gene sharing level values between the pithoviruses and cedratviruses was 42.1% and 24.8% respectively, and between the orpheovirus and the other viruses, the average was 32.1% and 8.2%, respectively (Fig. 2).
Phylogenomics
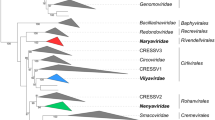
A concatenated phylogenetic approach based on seven core genes of giant viruses, named DNA polymerase family B, NCLDV major capsid protein, DEAD/SNF2-like helicase, DNA topoisomerase II, DNA-directed RNA polymerase beta subunit, transcription initiation factor IIB, and poxvirus late transcription factor VLTF3, was performed using representatives of the family Marseilleviridae as an outgroup. The analysis showed that cedratviruses are a sister group of pithoviruses with an equivalent phylogenetic breadth, and the orpheovirus forms a divergent branch with a wider breadth, supporting the creation of the three new families. The analysis was performed using only the isolates for which the whole genome sequence had been deposited in public databases at the time of the study, but complete high-quality viral genome sequences from metagenomic studies can provide additional evolutionary context that could result in revisions of the taxonomy proposed here.
Maximum-likelihood-based phylogenetic tree of seven marker genes, constructed using IQ-TREE multicore version 2.2.0 [38]. The best-fit model was LG + F + I + G4, chosen according to the Bayesian information criterion (BIC) [39]. Branch support was computed using 1000 ultrafast bootstrap replicates. The phylogenetic tree was visualized and edited using iTOL [40]. The members of the three proposed new families are indicated by different colors
Demarcation criteria
We propose that isolates with pairwise ANI > 95% should be considered members of the same species. Therefore, in our proposal, the species “Alphacedratvirus franciense” includes two viral isolates: cedratvirus lausannensis and cedratvirus zaza. All other isolates included here should be classified in separate new species.
In consensus with the newly recognized family Mamonoviridae, we propose that representatives of the same genus within the three families proposed here should have a pairwise ANI > 70% and similar morphological traits. Thus, according to these criteria, each of the three families is monogenic [12].
Families should be monophyletic clades with a high level of bootstrap support. Inclusion in the families should be validated based on comparative genomic analysis similar to that presented in this study, as well as phylogenetic analysis based on the seven genes (DNA PolB, MCP, DEAD/SNF2-like, DNA topoII, RNAPII, TIFIIB, VLTF3) that have been identified as the best markers for phylogenetic reconstructions of giant viruses (Fig. 3) [41]. Future genomes can be included in the phylogenomic analysis even if they differ and lack a subset of these genes.
We acknowledge that the proposed demarcation criteria are adaptable and subject to future reassessment as new viruses are isolated and their complete genomes are sequenced.
Conclusion
The number of characterized giant viruses within the order Pimascovirales has increased over the last decade, indicating a vast diversity yet to be discovered [1,2,3, 17, 19,20,21, 23, 25,26,27,28,29,30,31,32,33,34,35]. However, a group of phylogenetically related and well-characterized giant viruses that were putative members of this order still lacked formal classification. This group comprises three well-established clades of isolated viruses, commonly referred to as “pithoviruses”, “cedratviruses”, and “orpheoviruses”. Despite their common features, these ovoid-shaped giant viruses exhibit significant differences in gene content, genomic architecture, and the phylogenetic history of several core genes [16]. Additionally, despite the morphological similarity of their virions (ovoid shape), these viruses have distinct delivery portal characteristics. Cedratviruses feature two corks at each end of the viral particle, pithoviruses more often have only one cork, and orpheovirus has an ostiole at the apex of its particle. Moreover, orpheovirus also infects a different host, Vermamoeba vermiformis, whereas cedratviruses and pithoviruses infect amoebae of the genus Acanthamoeba [1,2,3]. Considering the dissimilarities within this group, and in line with the demarcation criteria of recently approved new taxa of giant viruses, our phylogenomic data suggest the creation of three family-level taxa in the order Pimascovirales. Accordingly, we proposed the creation of the families “Orpheoviridae”, “Cedratviridae”, and “Pithoviridae” [11, 12], each composed of a single genus. ("Alphaorpheovirus”, “Alphacedratvirus”, and “Alphapithovirus”, respectively), to formally accommodate eight isolates classified into seven new species.
The official taxonomy of giant viruses is currently in a very early stage. Therefore, the taxonomic framework proposed here represents an initial attempt to classify some of these viruses and is surely destined for future changes as new representatives are described and their genomes sequenced. The formal taxonomic proposal for classification of cedrat- and pithoviruses as well as an orpheovirus was submitted to the ICTV in June 2023 and was approved by a ratification vote in April 2024.
Data availability
The datasets generated and/or analyzed in the current study are available from the corresponding author on reasonable request.
References
Andreani J, Khalil JYB, Baptiste E, Hasni I, Michelle C, Raoult D, Levasseur A, La Scola B (2018) Orpheovirus IHUMI-LCC2: A New Virus among the Giant Viruses. Front Microbiol 8:2643. https://doi.org/10.3389/fmicb.2017.02643PMID: 29403444; PMCID: PMC5786535
Legendre M, Bartoli J, Shmakova L et al (2014) Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology. Proc Natl Acad Sci U S A 111:4274–4279. https://doi.org/10.1073/pnas.1320670111
Andreani J, Aherfi S, Bou Khalil JY, Di Pinto F, Bitam I, Raoult D, Colson P, La Scola B (2016) Cedratvirus, a Double-Cork Structured Giant Virus, is a Distant Relative of Pithoviruses. Viruses 8(11):300. https://doi.org/10.3390/v8110300PMID: 27827884; PMCID: PMC5127014
Philippe N, Legendre M, Doutre G et al (2013) Pandoraviruses: Amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science (1979) 341:281–286. https://doi.org/10.1126/science.1239181
Legendre M, Lartigue A, Bertaux L et al (2015) In-depth study of Mollivirus sibericum, a new 30,000-y-old giant virus infecting Acanthamoeba. Proc Natl Acad Sci 112(E5327–E5335). https://doi.org/10.1073/PNAS.1510795112
Andreani J, Khalil JYB, Sevvana M, Benamar S, Di Pinto F, Bitam I, Colson P, Klose T, Rossmann MG, Raoult D, La Scola B (2017) Pacmanvirus, a New Giant Icosahedral Virus at the Crossroads between Asfarviridae and Faustoviruses. J Virol 91(14):e00212–e00217. https://doi.org/10.1128/JVI.00212-17PMID: 28446673; PMCID: PMC5487549
Bajrai LH, Benamar S, Azhar EI, Robert C, Levasseur A, Raoult D, La Scola B (2016) Kaumoebavirus, a New Virus That Clusters with Faustoviruses and Asfarviridae. Viruses 8(11):278. https://doi.org/10.3390/v8110278PMID: 27801826; PMCID: PMC5127008
Reteno DG, Benamar S, Khalil JB et al (2015) Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. J Virol 89:6585. https://doi.org/10.1128/JVI.00115-15
Yoshikawa G, Blanc-Mathieu R, Song C et al (2019) Medusavirus, a Novel Large DNA Virus Discovered from Hot Spring Water. J Virol 93:2130–2148. https://doi.org/10.1128/jvi.02130-18
Rolland C, Andreani J, Sahmi-Bounsiar D et al (2021) Clandestinovirus: A Giant Virus With Chromatin Proteins and a Potential to Manipulate the Cell Cycle of Its Host Vermamoeba vermiformis. Front Microbiol 0:2224. https://doi.org/10.3389/FMICB.2021.715608
Aylward FO, Abrahão JS, Brussaard CPD, Fischer MG, Moniruzzaman M, Ogata H, Suttle CA (2023) Taxonomic update for giant viruses in the order Imitervirales (phylum Nucleocytoviricota). Arch Virol 168(11):283. https://doi.org/10.1007/s00705-023-05906-3. PMID: 37904060
Zhang R, Takemura M, Murata K, Ogata H (2023) Mamonoviridae, a proposed new family of the phylum Nucleocytoviricota. Arch Virol 168:1–6. https://doi.org/10.1007/S00705-022-05633-1
de Miranda Boratto PV, Oliveira GP, Abrahão JS (2022) Yaraviridae: a proposed new family of viruses infecting Acanthamoeba castellanii. Arch Virol 167:711–715. https://doi.org/10.1007/S00705-021-05326-1
Rodrigues RAL, Mougari S, Colson P et al (2019) Tupanvirus, a new genus in the family Mimiviridae. Arch Virol 164:325–331. https://doi.org/10.1007/S00705-018-4067-4
Current ICTV Taxonomy Release | ICTV. https://ictv.global/taxonomy. Accessed 14 Jun 2023
Queiroz VF, Carvalho JVRP, de Souza FG et al (2023) Analysis of the Genomic Features and Evolutionary History of Pithovirus-Like Isolates Reveals Two Major Divergent Groups of Viruses. J Virol 97(7):e0041123. https://doi.org/10.1128/JVI.00411-23
Levasseur A, Andreani J, Delerce J et al (2016) Comparison of a Modern and Fossil Pithovirus Reveals Its Genetic Conservation and Evolution. Genome Biol Evol 8:2333–2339. https://doi.org/10.1093/gbe/evw153
Postler TS, Rubino L, Adriaenssens EM et al (2022) Guidance for creating individual and batch latinized binomial virus species names. J Gen Virol 103:001800. https://doi.org/10.1099/JGV.0.001800
Bertelli C, Mueller L, Thomas V, Pillonel T, Jacquier N, Greub G (2017) Cedratvirus lausannensis - digging into Pithoviridae diversity. Environ Microbiol 19(10):4022–4034. https://doi.org/10.1111/1462-2920.13813Epub 2017 Aug 14. PMID: 28618143
Rodrigues RAL, Andreani J, Andrade ACDSP, Machado TB, Abdi S, Levasseur A, Abrahão JS, La Scola B (2018) Morphologic and Genomic Analyses of New Isolates Reveal a Second Lineage of Cedratviruses. J Virol 92(13):e00372–e00318. https://doi.org/10.1128/JVI.00372-18PMID: 29695424; PMCID: PMC6002711
Jeudy S, Rigou S, Alempic JM et al (2020) The DNA methylation landscape of giant viruses. Nat Commun 2020 11(1 11):1–12. https://doi.org/10.1038/s41467-020-16414-2
Souza F, Rodrigues R, Reis E et al (2019) In-depth analysis of the replication cycle of Orpheovirus. Virol J 16:158. https://doi.org/10.1186/s12985-019-1268-8
Silva LKDS, Andrade ACDSP, Dornas FP, Rodrigues RAL, Arantes T, Kroon EG, Bonjardim CA, Abrahão JS (2018) Cedratvirus getuliensis replication cycle: an in-depth morphological analysis. Sci Rep 8(1):4000. https://doi.org/10.1038/s41598-018-22398-3PMID: 29507337; PMCID: PMC5838162
Okamoto K, Miyazaki N, Song C et al (2017) Structural variability and complexity of the giant Pithovirus sibericum particle revealed by high-voltage electron cryo-tomography and energy-filtered electron cryo-microscopy. Sci Rep 2017 7(1 7):1–12. https://doi.org/10.1038/s41598-017-13390-4
Alempic JM, Lartigue A, Goncharov AE et al (2023) An Update on Eukaryotic Viruses Revived from Ancient Permafrost. Viruses 2023, Vol 15, Page 564 15:564. https://doi.org/10.3390/V15020564
Hikida H, Okazaki Y, Zhang R et al (2023) A rapid genome-wide analysis of isolated giant viruses using MinION sequencing. Environ Microbiol 25:2621–2635. https://doi.org/10.1111/1462-2920.16476
Rigou S, Schmitt A, Alempic JM, Lartigue A, Vendloczki P, Abergel C, Claverie JM, Legendre M (2023) Pithoviruses Are Invaded by Repeats That Contribute to Their Evolution and Divergence from Cedratviruses. Mol Biol Evol 40(11):msad244. https://doi.org/10.1093/molbev/msad244PMID: 37950899; PMCID: PMC10664404
Machado TB, Picorelli ACR, de Azevedo BL et al (2023) Gene duplication as a major force driving the genome expansion in some giant viruses. J Virol 97. https://doi.org/10.1128/JVI.01309-23
Andrade ACDSP, Arantes TS, Rodrigues RAL, Machado TB, Dornas FP, Landell MF, Furst C, Borges LGA, Dutra LAL, Almeida G, Trindade GS, Bergier I, Abrahão W, Borges IA, Cortines JR, de Oliveira DB, Kroon EG, Abrahão JS (2018) Ubiquitous giants: a plethora of giant viruses found in Brazil and Antarctica. Virol J 15(1):22. https://doi.org/10.1186/s12985-018-0930-xPMID: 29368617; PMCID: PMC5784613
Boudjemaa H, Andreani J, Bitam I, Scola B, La (2020) Diversity of Amoeba-Associated Giant Viruses Isolated in Algeria. Diversity 2020, Vol 12, Page 215 12:215. https://doi.org/10.3390/D12060215
Kördel M, Svenda M, Reddy HKN et al (2021) Quantitative conversion of biomass in giant DNA virus infection. Scientific Reports 2021 11:1 11:1–12. https://doi.org/10.1038/s41598-021-83547-9
Rigou S, Santini S, Abergel C et al (2022) Past and present giant viruses diversity explored through permafrost metagenomics. Nature Communications 2022 13:1 13:1–13. https://doi.org/10.1038/s41467-022-33633-x
Schulz F, Abergel C, Woyke T (2022) Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat Rev Microbiol 20:721–736. https://doi.org/10.1038/S41579-022-00754-5
Bäckström D, Yutin N, Jørgensen SL et al (2019) Virus Genomes from Deep Sea Sediments Expand the Ocean Megavirome and Support Independent Origins of Viral Gigantism. https://doi.org/10.1128/MBIO.02497-18. mBio 10:
Endo H, Blanc-Mathieu R, Li Y et al (2020) Biogeography of marine giant viruses reveals their interplay with eukaryotes and ecological functions. Nat Ecol Evol 4:1639–1649. https://doi.org/10.1038/s41559-020-01288-w
Darling AE, Mau B, Perna NT (2010) progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement. PLoS ONE 5:e11147. https://doi.org/10.1371/JOURNAL.PONE.0011147
Rodriguez -RLM, Konstantinidis KT (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. https://doi.org/10.7287/PEERJ.PREPRINTS.1900V1
Minh BQ, Schmidt HA, Chernomor O et al (2020) IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 37:1530–1534. https://doi.org/10.1093/MOLBEV/MSAA015
Kalyaanamoorthy S, Minh BQ, Wong TKF et al (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 2017 14:6. https://doi.org/10.1038/nmeth.4285
Letunic I, Bork P, Gmbh BS (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 1–4. https://doi.org/10.1093/nar/gkab301
Aylward FO, Moniruzzaman M, Ha AD, Koonin EV (2021) A phylogenomic framework for charting the diversity and evolution of giant viruses. PLoS Biol 19. https://doi.org/10.1371/JOURNAL.PBIO.3001430
Acknowledgements
We would like to thank our colleagues from Laboratório de Vírus—UFMG for their technical support. We acknowledge the outstanding work carried out by various research groups in isolating and characterizing cedratviruses, pithoviruses, and orpheovirus. This taxonomic proposal serves as a tribute to their dedication in this field. We are open to discussing the taxonomy and nomenclature of this virus group and develo** enhanced versions of this proposal in collaboration with the ICTV. RALR and JSA are CNPq researchers. The formal taxonomic proposal has now been approved by the ICTV.
Funding
We thank Minas Gerais State Agency for Research and Development (FAPEMIG), National Council for Scientific and Technological Development (CNPq), Coordination of Superior Level Staff Improvement (CAPES), and Pro-Reitorias de Pesquisa and Pós-Graduação-UFMG for financial support and scholarships.
Author information
Authors and Affiliations
Contributions
VFQ conducted analyses and drafted the initial manuscript. RALR and JSA designed the study, supervised its execution, and contributed to the final version of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
The topic of this study is registered at the Brazilian National System for the Management of Genetic Heritage and Associated Traditional Knowledge, number A2291C9.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Sead Sabanadzovic
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Queiroz, V.F., Rodrigues, R.A.L. & Abrahão, J.S. A taxonomic proposal for cedratviruses, orpheoviruses, and pithoviruses. Arch Virol 169, 132 (2024). https://doi.org/10.1007/s00705-024-06055-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00705-024-06055-x