Abstract
Porcine deltacoronavirus (PDCoV) is an enteric virus that was first identified in 2012. Although PDCoV has been detected worldwide, there is little information about its circulation in western China. In this study, fecal samples were collected from piglets with watery diarrhea in western China between 2015 and 2018 for the detection of PDCoV. The positive rate was 29.9%. A PDCoV strain (CHN/CQ/BN23/2016, BN23) was isolated and selected for further investigation. Phylogenetic analysis showed that this strain formed an individual cluster between the early Chinese lineage and the Chinese lineage. RDP4 and SimPlot analysis demonstrated that strain BN23 is a recombinant of Thailand/S5015L/2015 and CHN-AH-2004. The pathogenicity of BN23 was evaluated in 3-day-old piglets. Challenged piglets developed serious clinical signs and died at 3 days post-inoculation. Our data show that PDCoV is prevalent in western China and that strain BN23 is highly pathogenic to newborn piglets. Therefore, more attention should be paid to emerging PDCoV strains in western China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Porcine deltacoronavirus (PDCoV) is an emerging coronavirus that can cause enteric disease characterized by watery diarrhea, vomiting, dehydration, and growth retardation. The mortality rate is about 40%-80% in nursing piglets [1]. PDCoV is an enveloped positive-sense single-stranded RNA virus that belongs to the genus Deltacoronavirus within the family Coronaviridae [2]. The size of PDCoV genome is about 25.4 kb, the smallest genome among the known coronavirus. The genome encodes two large polyproteins (ORF1a and ORF1b), four structure proteins (spike, membrane, envelope, and nucleocapsid), and two accessory proteins (NS6 and NS7) [3, 4]. The spike (S) protein is the key responsible for virus entry. It also functions as the main antigen for the induction of protective antibodies. In addition, the S gene is the primary gene used for studying the genetic diversity of coronavirus isolates.
PDCoV was first reported in Hong Kong in 2012 [5]. Later, it caused outbreaks in the United States in 2014 [6]. Since then, the virus has spread to various countries, including Canada [7], South Korea [8], Japan [9], Vietnam [10], and Thailand [11, 12], causing tremendous financial losses to the pork industry.
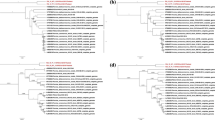
In China, the emergence of PDCoV was first reported in 2015 [13, 14]. Several PDCoV strains have been isolated in northern, southern, and central China [3A). To further characterize the putative recombination events, we performed nucleotide similarity comparisons between strain BN23 and other PDCoV strains, using SimPlot v.3.5.1. This analysis confirmed the BN23 was a recombinant strain with the recombination breakpoints map** to ORF1a (Fig. 3B). These results suggest that BN23 was generated by a natural recombination event between the Thailand/S5015L/2015 and CHN-AH-2004 strains (Tables 3, 4).
Recombination analysis of strain BN23. (A) Possible breakpoints in the recombination event involving strains Thailand/S5015L/2015 and CHN-AH-2004 were identified using the RDP method and confirmed using the RDP, GENECONV, Chimaera, Maxchi, BootScan, SiScan, and 3Seq applications in the RDP program (p < 0.01). (B) Nucleotide sequence similarity was assessed using SimPlot v.3.5.1.
CHN/CQ/BN23/2016 is highly pathogenic to newborn piglets
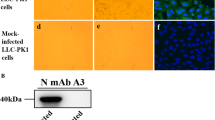
As PDCoV BN23 was found to be a recombinant strain that differs from the prevalent PDCoV strains, its pathogenicity was tested in piglets. Six 3-day-old piglets, free of PDCoV, PEDV, TGEV, PRV, and PCV-2, were divided randomly into two groups and inoculated with PDCoV BN23 (106 TCID50 per pig) or PBS. At one day postinfection (dpi), piglets inoculated with BN23 exhibited a loss of appetite. They also had mild diarrhea, and their body temperature decreased from about 39°C to 37°C. The piglets in the mock-infected group did not show clinical signs. At 2 dpi, the inoculated piglets developed severe diarrhea and consumed almost no milk, and their temperatures dropped to about 36 °C. All of the challenged piglets died at 3 dpi. The small intestines were found to be transparent, thin-walled, gas-distended, and filled with yellow watery content (Fig. 4A). No clinical signs were observed in the mock-infected piglets. Different tissues were also harvested for viral load determination, and the ileum was also fixed and subjected to histological analysis, which showed that the small-intestinal villi were mildly atrophied and contained aggregates of inflammatory cells. No lesions were observed in the mock-infected group (Fig. 4B). IFA analysis performed with sections showed that BN23 was present in the villus of the intestine (Fig. 4C). Real-time PCR analysis demonstrated that PDCoV BN23 has broad tissue tropism, with the viral genome detected in the duodenum, jejunum, ileum, colon, cecum, rectum, gastric mucosa, and mesenteric glands, with especially high viral loads in intestines. Within the intestines, higher viral copy numbers were found in the colon, cecum, and rectum than in the duodenum, jejunum, and ileum (Fig. 4D).
Pathogenicity evaluation in BN23-infected piglets. (A) Macroscopic lesions in BN23-challenged piglets and mock-infected piglets at 3 dpi. (B) Histologic lesions in the intestine caused by PDCoV CHN/CQ/BN23/2016. (C) IFA analysis of ileum sections. (D) Virus load in different tissues of piglets challenged with BN23. (D) Immune responses induced by PDCoV CHN/CQ/BN23/2016 infection in different segments of the intestine
We then examined whether PDCoV infection could induce enteric immunity. The results demonstrated that the IFN-λ3 gene was significantly upregulated in PDCoV-infected small intestines. Also, PDCoV infection enhanced the expression of IL-1β, IL-12A, and GM-CSF, suggesting that BN23 could induce inflammatory reactions in the intestine (Fig. 4E).
Discussion
PDCoV is a novel porcine enteric coronavirus. The clinical signs, including watery diarrhea, vomiting, and dehydration, are similar to those caused by PEDV [31]. In 2012, two PDCoV strains were first identified in Hong Kong [5]. Since then, PDCoV has been reported in other provinces of China [13, 14, 31, 36]. A retrospective study showed that PDCoV could be detected in samples from as early as 2004. A total of 215 samples collected during 2004–2014 in Anhui, Guangxi, Hubei, and Jiangsu provinces displayed a positive rate of 6.51% for PDCoV [36]. In previous studies, the prevalence of PDCoV infection was 23.4% in samples collected from Shanxi, Guangdong, and Hubei province, and it was 33.71% (120/356) in Jiangxi provinces since 2014 [13, 31]. Those results showed that PDCoV is circulating in southern China, but few studies on PDCoV have been performed in western China. Therefore, samples from piglets with watery diarrhea in western China were collected to test for PDCoV. The results demonstrated that PDCoV strains are also highly prevalent in western China, and one PDCoV strain was later isolated.
Phylogenetic analysis was performed to examine the evolutionary history of the new PDCoV isolate CHN/CQ/BN23/2016. Its complete genome showed 97.5%-99.8% sequence identity with other Chinese strains at the nucleotide level (Supplementary Table S2). It formed an individual subcluster between the Chinese lineage and the early Chinese lineage. The early Chinese lineage also included several strains isolated in Gansu and Qinhai provinces since 2016 [37]. These two areas were also invested in this study. The frequent identification of early Chinese strains suggested that the early Chinese PDCoV strains might have circulated in western China and recombined with PDCoV strains of other lineages. The phylogenetic analysis also revealed that the 3’UTR of BN23 belongs to the United States lineage, whereas the 5’UTR belongs to the early Chinese lineage. Recombination analysis indicated that a recombination event had occurred within ORF1a of BN23. The major parent strain of BN23 was CHN-AH-2004, while the minor parent was Thailand/S5015L/2015. In western China, Sichuan province contributes greatly to the pig production industry. The five provinces investigated in this study are not the traditional pig husbandry areas. Piglets are usually transported from Sichuan to the other five provinces. Moreover, Sichuan is an important transport hub for both western China and Southeast Asia. The frequent transport of animals may result in the emergence of recombinant PDCoV strains.
An animal experiment showed that BN23 is highly pathogenic to newborn piglets. The lamina propria was heavily infiltrated by inflammatory cells such as macrophages, lymphocytes, neutrophils, and eosinophils, which is in agreement with previous studies [24, 38, 39]. However, recently isolated Chinese PDCoV strains belonging to the Chinese lineage have been found to cause much milder disease in sucking piglets. In those studies, although typical symptoms were observed in infected piglets, most recovered from the infection [40,41,42]. Our results suggest that BN23 is more pathogenic than with other PDCoV strains belonging to the Chinese lineage. Moreover, BN23 infection elevated the expression of IFN-λ3, which results in a powerful response against the infection within the intestine. Inflammatory cytokines, including IL-1β, IL-12A, and GM-CSF, were also upregulated during BN23 infection. Inflammatory reactions were also observed in previous studies and correlated with H&E straining, suggesting that PDCoV infection causes a strong immune response [43]. Although it has been reported that PDCoV infection inhibits IFN-associated response in vivo [44], the opposite phenomenon was observed in vitro. The complex microenvironment within the intestine may be responsible for this difference.
In summary, this study demonstrated that PDCoV is prevalent in western China, and a new PDCoV strain, CHN/CQ/BN23/2016, showing genetic divergence from other Chinese strains, was identified. This virus formed an individual cluster between the early Chinese lineage and the Chinese lineage and was found to be a recombinant strain. It was also found to be highly pathogenic to newborn piglets. These results provide important information on the evolution of PDCoV in western of China and suggest that more surveillance is needed.
Abbreviations
- PDCoV:
-
Porcine deltacoronavirus
- PEDV:
-
Porcine epidemic diarrhea virus
- TGEV:
-
Transmissible gastroenteritis coronavirus
- PRV:
-
Pseudorabies
- PCV-2:
-
Porcine circovirus type 2
- UTR:
-
Untranslated region
- ORF:
-
Open reading frame
- PCR:
-
Polymerase chain reaction
- PBS:
-
Phosphate-buffered saline
- DPI:
-
Days postinfection
- HE:
-
Hematoxylin and eosin
- CPE:
-
Cytopathic effect
- RDP4:
-
Recombination Detection Program version.4.9.4
- IFA:
-
Immunofluorescence assay
References
Saeng-Chuto K, Jermsutjarit P, Stott CJ, Vui DT, Tantituvanont A, Nilubol D (2020) Retrospective study, full-length genome characterization and evaluation of viral infectivity and pathogenicity of chimeric porcine deltacoronavirus detected in Vietnam. Transbound Emerg Dis 67:183–198
Vitosh-Sillman S, Loy JD, Brodersen B, Kelling C, Doster A, Topliff C, Nelson E, Bai J, Schirtzinger E, Poulsen E, Meadors B, Anderson J, Hause B, Anderson G, Hesse R (2016) Experimental infection of conventional nursing pigs and their dams with Porcine deltacoronavirus. J Vet Diagnostic Investig 28:486–497
Lee S, Lee C (2014) Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announcements 2
Li G, Chen Q, Harmon KM, Yoon KJ, Schwartz KJ, Hoogland MJ, Gauger PC, Main RG, Zhang J (2014) Full-length genome sequence of porcine deltacoronavirus strain USA/IA/2014/8734. Genome Announcements 2
Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY (2012) Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol 86:3995–4008
Wang L, Byrum B, Zhang Y (2014) Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227–1230
Niederwerder MC, Hesse RA (2018) Swine enteric coronavirus disease: a review of 4 years with porcine epidemic diarrhoea virus and porcine deltacoronavirus in the United States and Canada. Transbound Emerg Dis 65:660–675
Lee JH, Chung HC, Nguyen VG, Moon HJ, Kim HK, Park SJ, Lee CH, Lee GE, Park BK (2016) Detection and phylogenetic analysis of porcine deltacoronavirus in Korean Swine Farms, 2015. Transbound Emerg Dis 63:248–252
Suzuki T, Shibahara T, Imai N, Yamamoto T, Ohashi S (2018) Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect Genet Evolut 61:176–182
Le VP, Song S, An BH, Park GN, Pham NT, Le DQ, Nguyen VT, Vu TTH, Kim KS, Choe S, An DJ (2018) A novel strain of porcine deltacoronavirus in Vietnam. Arch Virol 163:203–207
Janetanakit T, Lumyai M, Bunpapong N, Boonyapisitsopa S, Chaiyawong S, Nonthabenjawan N, Kesdaengsakonwut S, Amonsin A (2016) Porcine deltacoronavirus, Thailand, 2015. Emerg Infect Dis 22:757–759
Lorsirigool A, Saeng-Chuto K, Madapong A, Temeeyasen G, Tripipat T, Kaewprommal P, Tantituvanont A, Piriyapongsa J, Nilubol D (2017) The genetic diversity and complete genome analysis of two novel porcine deltacoronavirus isolates in Thailand in 2015. Virus Genes 53:240–248
Chen F, Zhu Y, Wu M, Ku X, Yao L, He Q (2015) Full-length genome characterization of Chinese porcine deltacoronavirus strain CH/SXD1/2015. Genome announcements 3
Wang YW, Yue H, Fang W, Huang YW (2015) Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announcements 3
Dong N, Fang L, Yang H, Liu H, Du T, Fang P, Wang D, Chen H, **ao S (2016) Isolation, genomic characterization, and pathogenicity of a Chinese porcine deltacoronavirus strain CHN-HN-2014. Vet Microbiol 196:98–106
Hsueh FC, Hsu FY, Chen YH, Shih HC, Lin WH, Yang CY, Lin CF, Chiou MT, Lin CN (2021) Phylogenetic classification of global porcine deltacoronavirus (PDCoV) reference strains and molecular characterization of PDCoV in Taiwan. Viruses 13
Huang H, Li Y, Wang W, Zheng M, Cao L, Sun W, Lu H (2020) Detection and molecular characterization of novel porcine bufaviruses in Guangxi province. Infect Genet Evolut 82:104286
Huang H, Yin Y, Wang W, Cao L, Sun W, Shi K, Lu H, ** N (2020) Emergence of Thailand-like strains of porcine deltacoronavirus in Guangxi Province, China. Vet Med Sci 6:854–859
** XH, Zhang YF, Yuan YX, Han L, Zhang GP, Hu H (2021) Isolation, characterization and transcriptome analysis of porcine deltacoronavirus strain HNZK-02 from Henan Province, China. Mol Immunol 134:86–99
Liu BJ, Zuo YZ, Gu WY, Luo SX, Shi QK, Hou LS, Zhong F, Fan JH (2018) Isolation and phylogenetic analysis of porcine deltacoronavirus from pigs with diarrhoea in Hebei province, China. Transbound Emerg Dis 65:874–882
Ding G, Fu Y, Li B, Chen J, Wang J, Yin B, Sha W, Liu G (2020) Development of a multiplex RT-PCR for the detection of major diarrhoeal viruses in pig herds in China. Transbound Emerg Dis 67:678–685
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003
Martin D, Rybicki E (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics (Oxford, England) 16:562–563
Padidam M, Sawyer S, Fauquet CM (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265:218–225
Gibbs MJ, Armstrong JS, Gibbs AJ (2000) Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics (Oxford, England) 16:573–582
Smith JM (1992) Analyzing the mosaic structure of genes. J Mol Evol 34:126–129
Boni MF, Posada D, Feldman MW (2007) An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 176:1035–1047
Holmes EC, Worobey M, Rambaut A (1999) Phylogenetic evidence for recombination in dengue virus. Mol Biol Evol 16:405–409
Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y (2015) Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis 62:575–580
Zhai SL, Wei WK, Li XP, Wen XH, Zhou X, Zhang H, Lv DH, Li F, Wang D (2016) Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol J 13:136
Zhang F, Luo S, Gu J, Li Z, Li K, Yuan W, Ye Y, Li H, Ding Z, Song D, Tang Y (2019) Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet Res 15:470
Zhang H, Liang Q, Li B, Cui X, Wei X, Ding Q, Wang Y, Hu H (2019) Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan Province, China. Prev Vet Med 166:8–15
Kong F, Wang Q, Kenney SP, Jung K, Vlasova AN, Saif LJ (2022) Porcine deltacoronaviruses: origin, evolution, cross-species transmission and zoonotic potential. Pathogens (Basel, Switzerland) 11
Dong N, Fang L, Zeng S, Sun Q, Chen H, **ao S (2015) Porcine deltacoronavirus in mainland China. Emerg Infect Dis 21:2254–2255
Wang M, Wang Y, Baloch AR, Pan Y, Tian L, Xu F, Shivaramu S, Chen S, Zeng Q (2018) Detection and genetic characterization of porcine deltacoronavirus in Tibetan pigs surrounding the Qinghai-Tibet Plateau of China. Transbound Emerg Dis 65:363–369
Chen Q, Gauger P, Stafne M, Thomas J, Arruda P, Burrough E, Madson D, Brodie J, Magstadt D, Derscheid R, Welch M, Zhang J (2015) Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 482:51–59
Wang L, Hayes J, Sarver C, Byrum B, Zhang Y (2016) Porcine deltacoronavirus: histological lesions and genetic characterization. Arch Virol 161:171–175
Li J, Zhou J, Zhao S, Guo R, Zhong C, Xue T, Peng Q, Zhang B, Fan B, Liu C, Ni Y, Ren L, Zhu X, Li B (2022) Pathogenicity, infective dose and altered gut microbiota in piglets infected with porcine deltacoronavirus. Virology 567:26–33
Wang H, Qin Y, Zhao W, Yuan T, Yang C, Mi X, Zhao P, Lu Y, Lu B, Chen Z, He Y, Yang C, Yi X, Wu Z, Chen Y, Wei Z, Huang W, Ouyang K (2021) Genetic characteristics and pathogenicity of a novel porcine deltacoronavirus Southeast Asia-Like Strain Found in China. Front Vet Sci 8:701612
Zhou X, Zhou L, Zhang P, Ge X, Guo X, Han J, Zhang Y, Yang H (2021) A strain of porcine deltacoronavirus: genomic characterization, pathogenicity and its full-length cDNA infectious clone. Transbound Emerg Dis 68:2130–2146
Zhang Q, Yoo D (2016) Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res 226:128–141
Zhang K, Lin S, Li J, Deng S, Zhang J, Wang S (2022) Modulation of innate antiviral immune response by porcine enteric coronavirus. Front Microbiol 13:845137
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31702209).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Handling Editor: Sheela Ramamoorthy.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Z., Li, S., Shao, Y. et al. Genomic characterization and pathogenicity analysis of a porcine deltacoronavirus strain isolated in western China. Arch Virol 167, 2249–2262 (2022). https://doi.org/10.1007/s00705-022-05549-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-022-05549-w