Abstract
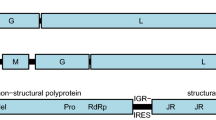
We report two novel RNA viruses from yellow crazy ants, (Anoplolepis gracilipes) detected using next-generation sequencing. The complete genome sequences of the two viruses were 10,662 and 8,238 nucleotides in length, respectively, with both possessing two open reading frames with three conserved protein domains. The genome organization is characteristic of members of the genus Triatovirus in the family Dicistroviridae. The two novel viruses were tentatively named “Anoplolepis gracilipes virus 1” and “Anoplolepis gracilipes virus 2” (AgrV-1 and AgrV-2). Phylogenetic analyses based on amino acid sequences of the non-structural polyprotein (ORF1) suggest that the two viruses are triatovirus-like viruses. This is the first report on the discovery of novel triatovirus-like viruses in yellow crazy ants with a description of their genome structure (two ORFs and conserved domains of RNA helicase, RNA-dependent RNA polymerase, and capsid protein), complete sequences, and viral prevalence across the Asia-Pacific region.


Similar content being viewed by others
Data availability
All viral sequences in this article are publicly available (GenBank accession numbers: MT108239 and MT108240) and also available as supplementary materials.
Change history
02 September 2020
Authors would like to correct the error in Fig. 1 which was incorrectly updated in the original publication.
References
Valles SM, Chen Y, Firth AE, Guérin DM, Hashimoto Y, Herrero S, de Miranda JR, Ryabov E (2017) ICTV virus taxonomy profile: dicistroviridae. J Gen Virol 98:355–356
Kleanthous E, Olendraite I, Lukhovitskaya NI, Firth AE (2019) Discovery of three RNA viruses using ant transcriptomic datasets. Arch Virol 164:643–647
Valles SM, Rivers AR (2019) Nine new RNA viruses associated with the fire ant Solenopsis invicta from its native range. Virus Genes 55:368–380
Viljakainen L, Holmberg I, Abril S, Jurvansuu J (2018) Viruses of invasive Argentine ants from the European Main supercolony: characterization, interactions and evolution. J Gen Virol 99:1129–1140
Reuter G, Pankovics P, Gyöngyi Z, Delwart E, Boros Á (2014) Novel dicistrovirus from bat guano. Arch Virol 159:3453–3456
Shi M, Lin XD, Tian JH, Chen LJ, Chen X, Li CX, Qin XC, Li J, Cao JP, Eden JS, Buchmann J (2016) Redefining the invertebrate RNA virosphere. Nature 540:539–543
Mihara T, Nishimura Y, Shimizu Y, Nishiyama H, Yoshikawa G, Uehara H, Hingamp P, Goto S, Ogata H (2016) Linking virus genomes with host taxonomy. Viruses 8:66
Payne AN, Shepherd TF, Rangel J (2020) The detection of honey bee (Apis mellifera) associated viruses in ants. Sci Rep 10:2923
Cooling M, Gruber MAM, Hoffmann BD, Sébastien A, Lester PJ (2017) A metatranscriptomic survey of the invasive yellow crazy ant, Anoplolepis gracilipes, identifies several potential viral and bacterial pathogens and mutualists. Insect Soc 64:197–207
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30:2114–2120
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29:644–652
Buchfink B, **e C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60
Huson D, Beier S, Flade I, Gorska A, El-Hadidi M, Mitra S, Ruscheweyh H, Rewati Tappu D (2016) MEGAN community edition—interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol 12:e1004957
Rombel IT, Sykes KF, Rayner S, Johnston SA (2002) ORF-FINDER: a vector for high-throughput gene identification. Gene 282:33–41
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226
Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T (2020) ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol 37:291–294
Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35:4453–4455
Letunic I, Bork P (2019) Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Yang CC, Yu YC, Valles SM, Oi DH, Chen YC, Shoemaker DW, Wu WJ, Shih CJ (2010) Loss of microbial (pathogen) infections associated with recent invasions of the red imported fire ant Solenopsis invicta. Biol Invasions 12:3307–3318
Culley AIL, Suttle CA (2012) Family dicistroviridae. In: King AMQA, Carstens EB, Lefkowitz EJ (eds) Virus taxonomy, classification and nomenclature of viruses. 9th report of the ICTV Elsevier Academic Press, Amsterdam, pp 840–854
Acknowledgements
We would like to express our sincere gratitude to the Research Institute for Sustainable Humanosphere, Kyoto University and the Environment Research and Ministry of the Environment, Japan, for financial support. We also extend our appreciation to Kazuki Tsuji, Chung-Chi Lin, Chow-Yang Lee, Ching-Chen Lee, Hui-Siang Tee, Mark Ooi, and Zhengwei Jong for field assistance, as well as three anonymous reviewers for constructive comments on earlier versions of the manuscript.
Funding
This research was financially supported by the Research Institute for Sustainable Humanosphere, Kyoto University and the Environment Research and Technology Development Fund, the Ministry of the Environment, Japan.
Author information
Authors and Affiliations
Contributions
C-CL: virus genome assembly, annotation, phylogenetic analysis, manuscript preparation. C-YL: RNA preparation, high throughput sequencing, manuscript preparation. H-WH: primer design, RT-PCR, and wet lab work. C-CSY: supervising all work, providing advice, securing funding, manuscript preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Ana Cristina Bratanich.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: Correct version of figure 1 updated here.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lee, CC., Lin, CY., Hsu, HW. et al. Complete genome sequences of two novel dicistroviruses detected in yellow crazy ants (Anoplolepis gracilipes). Arch Virol 165, 2715–2719 (2020). https://doi.org/10.1007/s00705-020-04769-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-020-04769-2