Abstract
Despite the growing knowledge of the clinicopathological features of COVID-19, the correlation between early changes in the laboratory parameters and the clinical outcomes of patients is not entirely understood. In this study, we aimed to assess the prognostic value of early laboratory parameters in COVID-19. We conducted a systematic review and meta-analysis based on the available literature in five databases. The last search was on July 26, 2020, with key terms related to COVID-19. Eligible studies contained original data of at least ten infected patients and reported on baseline laboratory parameters of patients. We calculated weighted mean differences (WMDs) for continuous outcomes and odds ratios (ORs) with 95% confidence intervals. 93 and 78 studies were included in quantitative and qualitative syntheses, respectively. Higher baseline total white blood cell count (WBC), C-reactive protein (CRP), lactate-dehydrogenase (LDH), creatine kinase (CK), D-dimer and lower absolute lymphocyte count (ALC) (WMDALC = − 0.35 × 109/L [CI − 0.43, − 0.27], p < 0.001, I2 = 94.2%; < 0.8 × 109/L, ORALC = 3.74 [CI 1.77, 7.92], p = 0.001, I2 = 65.5%) were all associated with higher mortality rate. On admission WBC, ALC, D-dimer, CRP, LDH, and CK changes could serve as alarming prognostic factors. The correct interpretation of laboratory abnormalities can guide therapeutic decisions, especially in early identification of potentially critical cases. This meta-analysis should help to allocate resources and save lives by enabling timely intervention.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Coronavirus disease-19 (COVID-19) is a novel coronavirus infection caused by the novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), which was first detected in Wuhan, China, in December 2019 after a series of pneumonia cases of unknown aetiology had emerged [1]. On 11 March 2020, WHO declared the rapid spread of this virus a pandemic [2]. Since the initial detection of the virus, more than 25,000,000 cases of COVID-19 have been confirmed worldwide with over 850,000 fatal cases [3].
In some patients, symptoms of severe respiratory infection can occur with rapidly develo** acute respiratory distress syndrome and other serious complications, which may be followed eventually by multiple organ failure and death. Therefore, early diagnosis and timely treatment of critical cases are crucial.
Despite some knowledge of the clinicopathological features of COVID-19, the correlation of changes in laboratory parameters and the prognosis of patients with COVID-19 is still unclear. However, studies on COVID-19 cases have shown that increased levels of white blood cells (WBC), decreased numbers of lymphocytes, especially CD8 + cells, increased levels of lactate-dehydrogenase (LDH), creatine kinase (CK), C-reactive protein (CRP), D-dimer, and levels of pro-inflammatory cytokines are associated with more severe inflammation and extensive lung damage with higher rates of admission to intensive care unit (ICU) and mortality [4]. A better understanding of early prognostic clinical laboratory parameters could save many lives by enabling timely intervention and better resource allocation since ICU capacity is limited in most countries. In this meta-analysis, we aimed to explore the significance of changes in the laboratory parameters and assessed the correlation between clinical laboratory data and the clinical outcomes of patients with COVID-19.
Methods
This systematic review with meta-analysis is reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses Statement [5]. The review protocol was registered on PROSPERO (CRD42020176836).
Search strategy
The systematic literature search was conducted in MEDLINE (via PubMed), Embase, Cochrane Library (CENTRAL), Scopus, and Web of Science for studies published from 1st January 2020 to 9th April 2020. The following search terms were used: (”covid 19”) OR (“Wuhan virus”) OR (“coronavirus”) OR (“2019 nCoV”) OR (“SARS-cov-2”). There was no restriction on the language of the records.
Selection and eligibility criteria
We selected clinical studies reporting on at least ten confirmed SARS-CoV-2 infected patients (based on the WHO case definition) and their laboratory findings. Studies were included in the systematic review of data on at least one of the following variables could be extracted: total white blood cell count (WBC), absolute lymphocyte count (ALC), absolute neutrophil count (ANC), platelet count, absolute basophil count, absolute eosinophil count (AEC), absolute monocyte count (AMC), C-reactive protein (CRP), haemoglobin, ferritin, lactate dehydrogenase (LDH), creatine kinase (CK), procalcitonin (PCT), fibrinogen, D-dimer, and any interleukins or lymphocyte subsets (CD3 + , CD4 + , CD8 +). The titles, abstracts, and full texts of the studies were screened by four independent review authors in pairs based on predefined criteria. The decision to include a study in the meta-analysis was based upon the assessment of the two reviewers and, if necessary, by a third reviewer for the resolution of any disagreements. Reference lists in the included studies and reviews on this topic were searched for additional studies. Publications citing the included studies were screened in the Google Scholar academic search engine too. Those studies that had either proven or suspected overlap** populations were included only in the systematic review part of this paper. To clarify these overlaps, we tried to contact the corresponding authors. Studies with more than 10% unclosed cases were excluded.
Data extraction
Four review authors independently extracted data into a standardized data collection form. The following data were extracted from each eligible article: first and second author, publication year, study site, study design, gender, age, and the means, standard deviations, medians, ranges, and interquartile ranges (IQR) of the laboratory values and specific thresholds with the corresponding intensive care requirement and mortality ratio. Data extraction was validated by a fifth review author. Discrepancies were resolved by a third party.
Risk of bias assessment
Based on the recommendation of the Cochrane Prognosis Methods Group, the QUIPS tool was applied by two independent authors for assessing the risk of bias in the studies included. Any disagreement was resolved based on consensus [6].
Statistical analysis
Pooled mean difference (weighted mean difference, WMD) was calculated for continuous outcomes and pooled odds ratios (ORs) were calculated for dichotomous outcomes. Random effect model was applied to all of the analyses with DerSimonien-Laird estimation. Statistical heterogeneity was analysed using the I2 the χ2 tests to obtain probability values: p < 0.01 was defined as indicating significant heterogeneity. Where mean with standard deviation was not reported for any of the outcomes, they were estimated from median, interquartiles and range using the method of Wan (2014) [7]. We performed separate analyses for mortality based on the clinical characteristics of the study population: one for all hospitalized COVID-19 patients (the “mixed” population) and the other for only critically ill COVID-19 patients. Small study effect was evaluated by visual assessment of funnel plot asymmetry and by Egger’s test were more than ten studies where available. Statistical analyses were performed with Stata 15 SE (Stata Corp). In the case of potentially overlap** study populations, data from the study with higher participant numbers were used for each outcome. ORs were calculated where raw data were available, however, only those meta-analyses were interpreted where at least three non-overlap** studies were available, as required.
Results
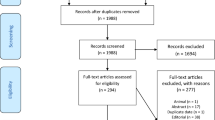
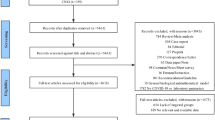
The results of our search and selection are detailed in the PRISMA-Flowchart shown in Fig. 1. Our systematic search yielded 93 eligible studies from 16 countries. We summarize the characteristics of the included studies in Supplementary Table 1. Out of these, fifty-six studies reported on the association of laboratory parameters and mortality. [8,9,10,11,12,13,14,15,16,17,18,33,34,35,36,37,38,39,Risk of bias assessment and publication bias Results of risk of bias assessments and evaluation of small-study effect are summarized in Supplementary Figures and among limitations of this study.
Discussion
In this meta-analysis, we have assessed the correlations between changes in laboratory parameters and the outcomes of patients with COVID-19. In doing so, we have identified many laboratory parameters that could be crucial for the timely identification of patients at higher risk of adverse outcomes.
This is the most comprehensive meta-analysis that assesses associations between on-admission laboratory parameters and mortality, as well as intensive care requirement. Compared with previous meta-analyses, [101,102,103,33, 130, 132].
We have noted that patients with a higher total WBC on admission had a poorer prognosis, while low total WBC levels were found to be a protective factor. Higher total WBC values are probably due mainly to increased levels of neutrophils [133]. In support of this idea, higher neutrophil counts also “predisposed” patients to unfavourable disease outcomes [134]. In light of our current knowledge, this might not be surprising since neutrophils are responsible for the production of pro-inflammatory mediators. Overproduction of these mediators, the so-called cytokine storm, has been suggested as a major cause of critical illness and mortality in COVID-19 [135].
It is important to note that increased levels of proinflammatory mediators such as CRP, fibrinogen and IL-6 were associated with worse outcomes. In agreement with previous studies, we found higher ferritin levels in non-survivors and critically ill patients. The laboratory profile in COVID-19 indicates hyperinflammation and may resemble secondary haemophagocytic lymphohistiocytosis (sHLH). However, other diagnostic criteria of sHLH have been rarely observed in COVID-19 [136,137,138].
This knowledge may help to identify therapeutic targets to minimize the cytokine storm. In addition, identifying those at higher risk of a cytokine storm is essential for treating them appropriately in advance [139].
Procalcitonin is not typically increased in viral infections; thus its elevated level at admission may not seem to be a significant finding in patients with COVID-19. Interestingly, according to our results, increased PCT levels have a predictive value for mortality, but not for intensive care requirement. An increase in its level might be associated with worse prognosis, possibly because of a bacterial superinfection, which could contribute to a rapid deterioration in the clinical course of disease towards multiorgan failure and death [140].
Compared to SARS-CoV, low platelet levels in COVID-19 are less common findings on admission. [141]. Although we found lower platelet levels in deceased patients compared to discharged ones, our pooled analyses did not indicate a clear prognostic role for platelet counts. However, studies found decreasing levels of platelet in patients are associated with adverse outcomes during the hospital stay [142, 143]. Thus, continuous monitoring of platelet counts may be required, even if its level initially gives no cause for concern.
Elevated D-dimer level is a typical sign of coagulation abnormalities in COVID-19 [144]. In our meta-analysis, increased D-dimer level was associated with worse prognosis in every comparison, except for the mean baseline D-dimer level between deceased and discharged critically ill patients (p = 0.149). However, the interpretation of these finding is uncertain since D-dimer levels can depend on several factors, including the presence of comorbidities or inflammatory processes [145].
The general indicators of tissue damage, elevated LDH and CK, were also associated with unfavourable outcomes in our meta-analysis, but none of these two laboratory parameters are specific for a special condition.
The underlying causes of the laboratory abnormalities are not entirely understood. Thus, further studies, including animal experiments, histological and pathological examinations, and clinical trials might give insight and identify potential therapeutic targets. More studies are required to further specify the thresholds applicable in clinical practice and resolve the contradiction in the role of certain biomarkers. Besides static values, the dynamics of laboratory parameters would worth further studying.
This meta-analysis has some limitations. Because of the nature of studies included, selection bias can occur, particularly in the case of parameters that are not routinely measured [62]. There was considerable heterogeneity in some analyses. Additionally, because of some studies with a high risk of bias, our results need to be interpreted cautiously. High risk of bias among studies mainly resulted from the significant differences in baseline characteristics of patients. Patients with advanced age and comorbidities are at higher risk both for more severe COVID-19 and for laboratory abnormalities. Conversion of medians to means could also distort our results. The visual assessment of funnel plots and Egger’s tests detected small-study effects in most of the analyses concerning WMD analyses.
In conclusion, we have shown that laboratory parameters on admission serve as important and early prognostic factors. These findings should help to allocate resources and potentially to save lives by enabling timely intervention.
Availability of data and material (data transparency)
The data that support the findings of this study are available from the corresponding author, [A.H.], upon reasonable request.
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus I, Research T (2020) A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 382(8):727–733. https://doi.org/10.1056/NEJMoa2001017
WHO (2020) WHO Director-General’s opening remarks at the media briefing on COVID-19—11 March 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-March-2020
Johns Hopkins University Coronavirus Resource Center. https://coronavirus.jhu.edu/. Accessed 30/08/2020
Du R-H, Liang L-R, Yang C-Q, Wang W, Cao T-Z, Li M, Guo G-Y, Du J, Zheng C-L, Zhu Q, Hu M, Li X-Y, Peng P, Shi H-Z (2020) Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. https://doi.org/10.1183/13993003.00524-2020
Moher D, Liberati A, Tetzlaff J, Altman DG, Group P (2009) Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339:b2535. https://doi.org/10.1136/bmj.b2535
Hayden JA, van der Wind DAT, Cartwright JL, Bombardier C (2013) Assessing bias in studies of prognostic factors. Ann Intern Med 158(4):280–286
Wan X, Wang W, Liu J, Tong T (2014) Estimating the sample mean and standard deviation from the sample size, median, range and/or interquartile range. BMC Med Res Methodol 14(1):135
Asghar MS, Haider Kazmi SJ, Ahmed Khan N, Akram M, Ahmed Khan S, Rasheed U, Hassan M, Memon GM (2020) Clinical profiles, characteristics, and outcomes of the first 100 admitted COVID-19 patients in pakistan: a single-center retrospective study in a Tertiary Care Hospital of Karachi. Cureus 12(6):e8712. https://doi.org/10.7759/cureus.8712
Auld SC, Caridi-Scheible M, Blum JM, Robichaux C, Kraft C, Jacob JT, Jabaley CS, Carpenter D, Kaplow R, Hernandez-Romieu AC, Adelman MW, Martin GS, Coopersmith CM, Murphy DJ, Clinical Research C (2020) ICU and ventilator mortality among critically Ill adults with coronavirus disease 2019. Crit Care Med 48(9):e799–e804. https://doi.org/10.1097/CCM.0000000000004457
Barman HA, Atici A, Sahin I, Alici G, Aktas Tekin E, Baycan OF, Ozturk F, Oflar E, Tugrul S, Yavuz MB, Celik FB, Oktay A, Vahaboglu H, Adas M, Turgut N, Okuyan E, Yildirmak MT, Gungor B (2020) Prognostic significance of cardiac injury in COVID-19 patients with and without coronary artery disease. Coron Artery Dis. https://doi.org/10.1097/MCA.0000000000000914
Bazzan M, Montaruli B, Sciascia S, Cosseddu D, Norbiato C, Roccatello D (2020) Low ADAMTS 13 plasma levels are predictors of mortality in COVID-19 patients. Intern Emerg Med 15(5):861–863. https://doi.org/10.1007/s11739-020-02394-0
Bhatraju PK, Ghassemieh BJ, Nichols M, Kim R, Jerome KR, Nalla AK, Greninger AL, Pipavath S, Wurfel MM, Evans L, Kritek PA, West TE, Luks A, Gerbino A, Dale CR, Goldman JD, O’Mahony S, Mikacenic C (2020) Covid-19 in critically Ill patients in the Seattle region—case series. N Engl J Med 382(21):2012–2022. https://doi.org/10.1056/NEJMoa2004500
Bonetti G, Manelli F, Patroni A, Bettinardi A, Borrelli G, Fiordalisi G, Marino A, Menolfi A, Saggini S, Volpi R, Anesi A, Lippi G (2020) Laboratory predictors of death from coronavirus disease 2019 (COVID-19) in the area of Valcamonica, Italy. Clin Chem Lab Med 58(7):1100–1105. https://doi.org/10.1515/cclm-2020-0459
Borobia AM, Carcas AJ, Arnalich F, Alvarez-Sala R, Monserrat-Villatoro J, Quintana M, Figueira JC, Torres Santos-Olmo RM, Garcia-Rodriguez J, Martin-Vega A, Buno A, Ramirez E, Martinez-Ales G, Garcia-Arenzana N, Nunez MC, Marti-de-Gracia M, Moreno Ramos F, Reinoso-Barbero F, Martin-Quiros A, Rivera Nunez A, Mingorance J, Carpio Segura CJ, Prieto Arribas D, Rey Cuevas E, Prados Sanchez C, Rios JJ, Hernan MA, Frias J, Arribas JR, On Behalf Of The Covid Hulp Working G (2020) A Cohort of Patients with COVID-19 in a Major Teaching Hospital in Europe. J Clin Med. https://doi.org/10.3390/jcm9061733
Cao J, Tu WJ, Cheng W, Yu L, Liu YK, Hu X, Liu Q (2020) Clinical features and short-term outcomes of 102 patients with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis 71(15):748–755. https://doi.org/10.1093/cid/ciaa243
Cen Y, Chen X, Shen Y, Zhang XH, Lei Y, Xu C, Jiang WR, Xu HT, Chen Y, Zhu J, Zhang LL, Liu YH (2020) Risk factors for disease progression in patients with mild to moderate coronavirus disease 2019-a multi-centre observational study. Clin Microbiol Infect. https://doi.org/10.1016/j.cmi.2020.05.041
Chen L, Yu J, He W, Chen L, Yuan G, Dong F, Chen W, Cao Y, Yang J, Cai L, Wu D, Ran Q, Li L, Liu Q, Ren W, Gao F, Wang H, Chen Z, Gale RP, Li Q, Hu Y (2020) Risk factors for death in 1859 subjects with COVID-19. Leukemia 34(8):2173–2183. https://doi.org/10.1038/s41375-020-0911-0
Chen R, Liang W, Jiang M, Guan W, Zhan C, Wang T, Tang C, Sang L, Liu J, Ni Z, Hu Y, Liu L, Shan H, Lei C, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Liu X, Cheng L, Ye F, Zheng J, Zhang N, Li Y, He J, Li S, Zhong N, Medical Treatment Expert Group for C (2020) Risk factors of fatal outcome in hospitalized subjects with Coronavirus Disease 2019 From a Nationwide Analysis in China. Chest 158 (1):97–105. https://doi.org/10.1016/j.chest.2020.04.010
Chen R, Sang L, Jiang M, Yang Z, Jia N, Fu W, **e J, Guan W, Liang W, Ni Z, Hu Y, Liu L, Shan H, Lei C, Peng Y, Wei L, Liu Y, Hu Y, Peng P, Wang J, Liu J, Chen Z, Li G, Zheng Z, Qiu S, Luo J, Ye C, Zhu S, Zheng J, Zhang N, Li Y, He J, Li J, Li S, Zhong N, Medical Treatment Expert Group for C (2020) Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J Allergy Clin Immunol 146(1):89–100. https://doi.org/10.1016/j.jaci.2020.05.003
Chen X, Zhao B, Qu Y, Chen Y, **ong J, Feng Y, Men D, Huang Q, Liu Y, Yang B, Ding J, Li F (2020) Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa449
Ciceri F, Castagna A, Rovere-Querini P, De Cobelli F, Ruggeri A, Galli L, Conte C, De Lorenzo R, Poli A, Ambrosio A, Signorelli C, Bossi E, Fazio M, Tresoldi C, Colombo S, Monti G, Fominskiy E, Franchini S, Spessot M, Martinenghi C, Carlucci M, Beretta L, Scandroglio AM, Clementi M, Locatelli M, Tresoldi M, Scarpellini P, Martino G, Bosi E, Dagna L, Lazzarin A, Landoni G, Zangrillo A (2020) Early predictors of clinical outcomes of COVID-19 outbreak in Milan, Italy. Clin Immunol 217:108509. https://doi.org/10.1016/j.clim.2020.108509
Cummings MJ, Baldwin MR, Abrams D, Jacobson SD, Meyer BJ, Balough EM, Aaron JG, Claassen J, Rabbani LE, Hastie J, Hochman BR, Salazar-Schicchi J, Yip NH, Brodie D, O’Donnell MR (2020) Epidemiology, clinical course, and outcomes of critically ill adults with COVID-19 in New York City: a prospective cohort study. The Lancet 395(10239):1763–1770. https://doi.org/10.1016/s0140-6736(20)31189-2
De Biasi S, Meschiari M, Gibellini L, Bellinazzi C, Borella R, Fidanza L, Gozzi L, Iannone A, Lo Tartaro D, Mattioli M, Paolini A, Menozzi M, Milic J, Franceschi G, Fantini R, Tonelli R, Sita M, Sarti M, Trenti T, Brugioni L, Cicchetti L, Facchinetti F, Pietrangelo A, Clini E, Girardis M, Guaraldi G, Mussini C, Cossarizza A (2020) Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun 11(1):3434. https://doi.org/10.1038/s41467-020-17292-4
Fan H, Zhang L, Huang B, Zhu M, Zhou Y, Zhang H, Tao X, Cheng S, Yu W, Zhu L, Chen J (2020) Cardiac injuries in patients with coronavirus disease 2019: not to be ignored. Int J Infect Dis 96:294–297. https://doi.org/10.1016/j.ijid.2020.05.024
Fan J, Wang H, Ye G, Cao X, Xu X, Tan W, Zhang Y (2020) Letter to the Editor: low-density lipoprotein is a potential predictor of poor prognosis in patients with coronavirus disease 2019. Metabolism 107:154243. https://doi.org/10.1016/j.metabol.2020.154243
Galloway JB, Norton S, Barker RD, Brookes A, Carey I, Clarke BD, **a R, Reid C, Russell MD, Sneep R, Sugarman L, Williams S, Yates M, Teo J, Shah AM, Cantle F (2020) A clinical risk score to identify patients with COVID-19 at high risk of critical care admission or death: an observational cohort study. J Infect 81(2):282–288. https://doi.org/10.1016/j.**f.2020.05.064
Gan J, Li J, Li S, Yang C (2020) Leucocyte subsets effectively predict the clinical outcome of patients with COVID-19 pneumonia: a retrospective case-control study. Front Public Health 8:299. https://doi.org/10.3389/fpubh.2020.00299
Giacomelli A, Ridolfo AL, Milazzo L, Oreni L, Bernacchia D, Siano M, Bonazzetti C, Covizzi A, Schiuma M, Passerini M, Piscaglia M, Coen M, Gubertini G, Rizzardini G, Cogliati C, Brambilla AM, Colombo R, Castelli A, Rech R, Riva A, Torre A, Meroni L, Rusconi S, Antinori S, Galli M (2020) 30-day mortality in patients hospitalized with COVID-19 during the first wave of the Italian epidemic: a prospective cohort study. Pharmacol Res 158:104931. https://doi.org/10.1016/j.phrs.2020.104931
Al-Samkari Hanny, Leaf Rebecca Karp, Dzik Walter, Carlson Jonathan, Fogerty Annemarie, Waheed Anem, Goodarzi Katayoon, Bendapudi Pavan, Bornikova Larissa, Gupta Shruti, Leaf David, Kuter David, Rosovsky R (2020) covid and coagulation: bleeding and thrombotic manifestations of SARS-COV2 infection. Blood 136(4):489–500. https://doi.org/10.1182/blood.2020006520/1743732/blood.2020006520.pdf
**ngwei He, **sheng Lai, Jia Cheng, Mengwen Wang, Yunjian Liu, **ao Zhichao Xu, Chang Li Shusheng, Hesong Z (2020) Impact of complicated myocardial injury on the clinical outcome of severe or critically ill COVID-19 patients. Chin J Cardiol 48:E011
Huang W, Li C, Wang Z, Wang H, Zhou N, Jiang J, Ni L, Zhang XA, Wang DW (2020) Decreased serum albumin level indicates poor prognosis of COVID-19 patients: hepatic injury analysis from 2,623 hospitalized cases. Sci China Life Sci. https://doi.org/10.1007/s11427-020-1733-4
Javanian M, Bayani M, Shokri M, Sadeghi-Haddad-Zavareh M, Babazadeh A, Yeganeh B, Mohseni S, Mehraeen R, Sepidarkish M, Bijani A, Rostami A, Shahbazi M, Tabari AM, Shabani A, Masrour-Roudsari J, Hasanpour AH, Gholinejad HE, Ghorbani H, Ebrahimpour S (2020) Clinical and laboratory findings from patients with COVID-19 pneumonia in Babol North of Iran: a retrospective cohort study. Rom J Intern Med 58(3):161–167. https://doi.org/10.2478/rjim-2020-0013
Li Juyi, Li Meng, Zheng Shasha, Li Menglan, Zhang Minghua, Sun Minxian, Li **, Cai Yi, Zhang H (2020) Plasma albumin levels predict risk for nonsurvivors in critically ill patients with COVID-19. Biomark Med. https://doi.org/10.2217/bmm-2020-0254
Li D, Chen Y, Liu H, Jia Y, Li F, Wang W, Wu J, Wan Z, Cao Y, Zeng R (2020) Immune dysfunction leads to mortality and organ injury in patients with COVID-19 in China: insights from ERS-COVID-19 study. Signal Transduct Target Ther 5(1):62. https://doi.org/10.1038/s41392-020-0163-5
Li K, Chen D, Chen S, Feng Y, Chang C, Wang Z, Wang N, Zhen G (2020) Predictors of fatality including radiographic findings in adults with COVID-19. Respir Res 21(1):146. https://doi.org/10.1186/s12931-020-01411-2
Li L, Yang L, Gui S, Pan F, Ye T, Liang B, Hu Y, Zheng C (2020) Association of clinical and radiographic findings with the outcomes of 93 patients with COVID-19 in Wuhan, China. Theranostics 10(14):6113–6121. https://doi.org/10.7150/thno.46569
Li Q, Cao Y, Chen L, Wu D, Yu J, Wang H, He W, Chen L, Dong F, Chen W, Chen W, Li L, Ran Q, Liu Q, Ren W, Gao F, Chen Z, Gale RP, Hu Y (2020) Hematological features of persons with COVID-19. Leukemia 34(8):2163–2172. https://doi.org/10.1038/s41375-020-0910-1
Li YK, Peng S, Li LQ, Wang Q, ** W, Zhang N, Fu XN (2020) Clinical and transmission characteristics of Covid-19—a retrospective study of 25 cases from a single thoracic surgery department. Curr Med Sci 40(2):295–300. https://doi.org/10.1007/s11596-020-2176-2
Liu Y, Sun W, Guo Y, Chen L, Zhang L, Zhao S, Long D, Yu L (2020) Association between platelet parameters and mortality in coronavirus disease 2019: retrospective cohort study. Platelets 31(4):490–496. https://doi.org/10.1080/09537104.2020.1754383
Long H, Nie L, **ang X, Li H, Zhang X, Fu X, Ren H, Liu W, Wang Q, Wu Q (2020) D-Dimer and Prothrombin Time Are the Significant Indicators of Severe COVID-19 and Poor Prognosis. Biomed Res Int 2020:1–10. https://doi.org/10.1155/2020/6159720
Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S (2020) IL-6 and CD8 + T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight. https://doi.org/10.1172/jci.insight.139024
Mikami T, Miyashita H, Yamada T, Harrington M, Steinberg D, Dunn A, Siau E (2020) Risk factors for mortality in patients with COVID-19 in New York City. J Gen Intern Med. https://doi.org/10.1007/s11606-020-05983-z
Omrani-Nava V, Maleki I, Ahmadi A, Moosazadeh M, Hedayatizadeh-Omran A, Roozbeh F, Nahanghi H, Alizadeh-Navaei R (2020) Evaluation of hepatic enzymes changes and association with prognosis in COVID-19 patients. Hepatitis Monthly. https://doi.org/10.5812/hepatmon.103179
Price-Haywood EG, Burton J, Fort D, Seoane L (2020) Hospitalization and mortality among black patients and white patients with Covid-19. N Engl J Med 382(26):2534–2543. https://doi.org/10.1056/NEJMsa2011686
Rivera-Izquierdo M, Del Carmen Valero-Ubierna M, Rd JL, Fernandez-Garcia MA, Martinez-Diz S, Tahery-Mahmoud A, Rodriguez-Camacho M, Gamiz-Molina AB, Barba-Gyengo N, Gamez-Baeza P, Cabrero-Rodriguez C, Guirado-Ruiz PA, Martin-Romero DT, Lainez-Ramos-Bossini AJ, Sanchez-Perez MR, Mancera-Romero J, Garcia-Martin M, Martin-delosReyes LM, Martinez-Ruiz V, Lardelli-Claret P, Jimenez-Mejias E (2020) Sociodemographic, clinical and laboratory factors on admission associated with COVID-19 mortality in hospitalized patients: a retrospective observational study. PLoS ONE 15(6):e0235107. https://doi.org/10.1371/journal.pone.0235107
Ruan Q, Yang K, Wang W, Jiang L, Song J (2020) Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 46(5):846–848. https://doi.org/10.1007/s00134-020-05991-x
Salacup G, Lo KB, Gul F, Peterson E, De Joy R, Bhargav R, Pelayo J, Albano J, Azmaiparashvili Z, Benzaquen S, Patarroyo-Aponte G, Rangaswami J (2020) Characteristics and clinical outcomes of COVID-19 patients in an underserved-inner city population: a single tertiary center cohort. J Med Virol. https://doi.org/10.1002/jmv.26252
Satici C, Demirkol MA, Sargin Altunok E, Gursoy B, Alkan M, Kamat S, Demirok B, Surmeli CD, Calik M, Cavus Z, Esatoglu SN (2020) Performance of pneumonia severity index and CURB-65 in predicting 30-day mortality in patients with COVID-19. Int J Infect Dis 98:84–89. https://doi.org/10.1016/j.ijid.2020.06.038
Shahriarirad R, Khodamoradi Z, Erfani A, Hosseinpour H, Ranjbar K, Emami Y, Mirahmadizadeh A, Lotfi M, Shirazi Yeganeh B, Dorrani Nejad A, Hemmati A, Ebrahimi M, Moghadami M (2020) Epidemiological and clinical features of 2019 novel coronavirus diseases (COVID-19) in the South of Iran. BMC Infect Dis 20(1):427. https://doi.org/10.1186/s12879-020-05128-x
Violi F, Cangemi R, Romiti GF, Ceccarelli G, Oliva A, Alessandri F, Pirro M, Pignatelli P, Lichtner M, Carraro A, Cipollone F, D’Ardes D, Pugliese F, Mastroianni CM (2020) Is albumin predictor of mortality in COVID-19? Antioxid Redox Signal. https://doi.org/10.1089/ars.2020.8142
Wang D, Yin Y, Hu C, Liu X, Zhang X, Zhou S, Jian M, Xu H, Prowle J, Hu B, Li Y, Peng Z (2020) Clinical course and outcome of 107 patients infected with the novel coronavirus, SARS-CoV-2, discharged from two hospitals in Wuhan, China. Crit Care 24(1):188. https://doi.org/10.1186/s13054-020-02895-6
Wang K, Zuo P, Liu Y, Zhang M, Zhao X, **e S, Zhang H, Chen X, Liu C (2020) Clinical and laboratory predictors of in-hospital mortality in patients with COVID-19: a cohort study in Wuhan. Clin Infect Dis, China. https://doi.org/10.1093/cid/ciaa538
Xu B, Fan CY, Wang AL, Zou YL, Yu YH, He C, **a WG, Zhang JX, Miao Q (2020) Suppressed T cell-mediated immunity in patients with COVID-19: a clinical retrospective study in Wuhan, China. J Infect 81(1):e51–e60. https://doi.org/10.1016/j.**f.2020.04.012
Xu J, Yang X, Yang L, Zou X, Wang Y, Wu Y, Zhou T, Yuan Y, Qi H, Fu S, Liu H, **a J, Xu Z, Yu Y, Li R, Ouyang Y, Wang R, Ren L, Hu Y, Xu D, Zhao X, Yuan S, Zhang D, Shang Y (2020) Clinical course and predictors of 60-day mortality in 239 critically ill patients with COVID-19: a multicenter retrospective study from Wuhan, China. Crit Care 24(1):394. https://doi.org/10.1186/s13054-020-03098-9
Hang Yang, Lin-cheng Yang, Rhui-tao Zhang, Yun-peng Ling, Qing-gang G (2020) Risk factors for death among COVID-19 patients combined qith hypertension, coronary heart disease or diabetes. J Peking Univ 180(7):1–11
Yao Q, Wang P, Wang X, Qie G, Meng M, Tong X, Bai X, Ding M, Liu W, Liu K, Chu Y (2020) A retrospective study of risk factors for severe acute respiratory syndrome coronavirus 2 infections in hospitalized adult patients. Pol Arch Intern Med 130(5):390–399. https://doi.org/10.20452/pamw.15312
Ye W, Chen G, Li X, Lan X, Ji C, Hou M, Zhang D, Zeng G, Wang Y, Xu C, Lu W, Cui R, Cai Y, Huang H, Yang L (2020) Dynamic changes of D-dimer and neutrophil-lymphocyte count ratio as prognostic biomarkers in COVID-19. Respir Res 21(1):169. https://doi.org/10.1186/s12931-020-01428-7
Yu C, Lei Q, Li W, Wang X, Liu W, Fan X, Li W (2020) Clinical characteristics, associated factors, and predicting COVID-19 mortality risk: a retrospective study in Wuhan, China. Am J Prev Med 59(2):168–175. https://doi.org/10.1016/j.amepre.2020.05.002
Zhang L, Yan X, Fan Q, Liu H, Liu X, Liu Z, Zhang Z (2020) D-dimer levels on admission to predict in-hospital mortality in patients with Covid-19. J Thromb Haemost 18(6):1324–1329. https://doi.org/10.1111/jth.14859
Zhao L, Zhang YP, Yang X, Liu X (2020) Eosinopenia is associated with greater severity in patients with coronavirus disease 2019. Allergy. https://doi.org/10.1111/all.14455
Zhao X, Wang K, Zuo P, Liu Y, Zhang M, **e S, Zhang H, Chen X, Liu C (2020) Early decrease in blood platelet count is associated with poor prognosis in COVID-19 patients-indications for predictive, preventive, and personalized medical approach. EPMA J. https://doi.org/10.1007/s13167-020-00208-z
Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, **ang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B (2020) Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet 395(10229):1054–1062. https://doi.org/10.1016/s0140-6736(20)30566-3
Zou X, Li S, Fang M, Hu M, Bian Y, Ling J, Yu S, **g L, Li D, Huang J (2020) Acute physiology and chronic health evaluation II Score as a Predictor of Hospital Mortality in Patients of Coronavirus Disease 2019. Crit Care Med 48(8):e657–e665. https://doi.org/10.1097/ccm.0000000000004411
Aggarwal S, Garcia-Telles N, Aggarwal G, Lavie C, Lippi G, Henry BM (2020) Clinical features, laboratory characteristics, and outcomes of patients hospitalized with coronavirus disease 2019 (COVID-19): early report from the United States. Diagnosis (Berl) 7(2):91–96. https://doi.org/10.1515/dx-2020-0046
Bhargava A, Fukushima EA, Levine M, Zhao W, Tanveer F, Szpunar SM, Saravolatz L (2020) Predictors for severe COVID-19 infection. Clin Infect Dis. https://doi.org/10.1093/cid/ciaa674
Burian E, Jungmann F, Kaissis GA, Lohofer FK, Spinner CD, Lahmer T, Treiber M, Dommasch M, Schneider G, Geisler F, Huber W, Protzer U, Schmid RM, Schwaiger M, Makowski MR, Braren RF (2020) Intensive care risk estimation in COVID-19 pneumonia based on clinical and imaging parameters: experiences from the Munich cohort. J Clin Med. https://doi.org/10.3390/jcm9051514
Cecconi M, Piovani D, Brunetta E, Aghemo A, Greco M, Ciccarelli M, Angelini C, Voza A, Omodei P, Vespa E, Pugliese N, Parigi TL, Folci M, Danese S, Bonovas S (2020) Early predictors of clinical deterioration in a cohort of 239 patients hospitalized for Covid-19 infection in Lombardy. J Clin Med, Italy. https://doi.org/10.3390/jcm9051548
Chan SSW, Christopher D, Tan GB, Chong VCL, Fan BE, Lin CY, Ong KH (2020) Peripheral lymphocyte subset alterations in COVID-19 patients. Int J Lab Hematol. https://doi.org/10.1111/ijlh.13276
Chen J, Qi T, Liu L, Ling Y, Qian Z, Li T, Li F, Xu Q, Zhang Y, Xu S, Song Z, Zeng Y, Shen Y, Shi Y, Zhu T, Lu H (2020) Clinical progression of patients with COVID-19 in Shanghai, China. J Infect 80(5):e1–e6. https://doi.org/10.1016/j.**f.2020.03.004
Cugno M, Meroni PL, Gualtierotti R, Griffini S, Grovetti E, Torri A, Panigada M, Aliberti S, Blasi F, Tedesco F, Peyvandi F (2020) Complement activation in patients with COVID-19: a novel therapeutic target. J Allergy Clin Immunol 146(1):215–217. https://doi.org/10.1016/j.jaci.2020.05.006
d’Alessandro M, Cameli P, Refini RM, Bergantini L, Alonzi V, Lanzarone N, Bennett D, Rana GD, Montagnani F, Scolletta S, Franchi F, Frediani B, Valente S, Mazzei MA, Bonella F, Bargagli E (2020) Serum KL-6 concentrations as a novel biomarker of severe COVID-19. J Med Virol. https://doi.org/10.1002/jmv.26087
Du RH, Liu LM, Yin W, Wang W, Guan LL, Yuan ML, Li YL, Hu Y, Li XY, Sun B, Peng P, Shi HZ (2020) Hospitalization and critical care of 109 decedents with COVID-19 pneumonia in Wuhan, China. Ann Am Thorac Soc 17(7):839–846. https://doi.org/10.1513/AnnalsATS.202003-225OC
Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, Mucheli SS, Kuperan P, Ong KH (2020) Hematologic parameters in patients with COVID-19 infection. Am J Hematol 95(6):E131–E134. https://doi.org/10.1002/ajh.25774
Feng Y, Ling Y, Bai T, **e Y, Huang J, Li J, **ong W, Yang D, Chen R, Lu F, Lu Y, Liu X, Chen Y, Li X, Li Y, Summah HD, Lin H, Yan J, Zhou M, Lu H, Qu J (2020) COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med 201(11):1380–1388. https://doi.org/10.1164/rccm.202002-0445OC
Cai SH, Liao W, Chen SW, Liu LL, Liu SY, Zheng ZD (2020) Association between obesity and clinical prognosis in patients infected with SARS-CoV-2. Infect Dis Poverty 9(1):80. https://doi.org/10.1186/s40249-020-00703-5
Goshua G, Pine AB, Meizlish ML, Chang CH, Zhang H, Bahel P, Baluha A, Bar N, Bona RD, Burns AJ, Dela Cruz CS, Dumont A, Halene S, Hwa J, Koff J, Menninger H, Neparidze N, Price C, Siner JM, Tormey C, Rinder HM, Chun HJ, Lee AI (2020) Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. The Lancet Haematol 7(8):e575–e582. https://doi.org/10.1016/s2352-3026(20)30216-7
Ihle-Hansen Håkon, Berge Trygve, Tveita Anders, Rønning Else Johanne, Ernø Per Erik, Andersen Elizabeth Lyster, Wang Christian Hjorth, Tveit Arnljot, Myrstad M (2020) Covid-19: symptomer, forløp og bruk av kliniske skåringsverktøy hos de 42 første pasientene innlagt på et norsk lokalsykehus. Tidsskrift for Den norske legeforening. https://doi.org/10.4045/tidsskr.20.0301
Hong KS, Lee KH, Chung JH, Shin KC, Choi EY, ** HJ, Jang JG, Lee W, Ahn JH (2020) Clinical features and outcomes of 98 patients hospitalized with SARS-CoV-2 infection in Daegu, South Korea: a brief descriptive study. Yonsei Med J 61(5):431–437. https://doi.org/10.3349/ymj.2020.61.5.431
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, **a J, Wei Y, Wu W, **e X, Yin W, Li H, Liu M, **ao Y, Gao H, Guo L, **e J, Wang G, Jiang R, Gao Z, ** Q, Wang J, Cao B (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395(10223):497–506. https://doi.org/10.1016/s0140-6736(20)30183-5
Jean-Luc Murk, Rik van de Biggelaar, Joep Stohr JV, Anton Buiting SW, Martijn van Hooft, Bram Diederen, Yvette Kluiters-de Hingh, Erik Ranschaert, Annemarie Brouwer, Jeroen Retera, Margot Verheijen, Dharmanand Ramnarain, Oers IvEeJv (2020) De eerste honderd opgenomen COVID-19-patiënten in het ElisabethTweesteden Ziekenhuis. Nederlands Tijdschrift voor Geneeskunde 164
Khamis F, Al-Zakwani I, Al Naamani H, Al Lawati S, Pandak N, Omar MB, Al Bahrani M, Bulushi ZA, Al Khalili H, Al Salmi I, Al Ismaili R, Al Awaidy ST (2020) Clinical characteristics and outcomes of the first 63 adult patients hospitalized with COVID-19: an experience from Oman. J Infect Public Health 13(7):906–913. https://doi.org/10.1016/j.jiph.2020.06.002
Lagi F, Piccica M, Graziani L, Vellere I, Botta A, Tilli M, Ottino L, Borchi B, Pozzi M, Bartalesi F, Mencarini J, Spinicci M, Zammarchi L, Pieralli F, Zagli G, Nozzoli C, Romagnoli S, Bartoloni A (2020) Early experience of an infectious and tropical diseases unit during the coronavirus disease (COVID-19) pandemic, Florence, Italy, February to March 2020. Eur Surveill. https://doi.org/10.2807/1560-7917.es.2020.25.17.2000556
Li H, **ang X, Ren H, Xu L, Zhao L, Chen X, Long H, Wang Q, Wu Q (2020) Serum Amyloid A is a biomarker of severe Coronavirus Disease and poor prognosis. J Infect 80(6):646–655. https://doi.org/10.1016/j.**f.2020.03.035
Liu R, Wang Y, Li J, Han H, **a Z, Liu F, Wu K, Yang L, Liu X, Zhu C (2020) Decreased T cell populations contribute to the increased severity of COVID-19. Clin Chim Acta 508:110–114. https://doi.org/10.1016/j.cca.2020.05.019
Liu Y, Yang Y, Zhang C, Huang F, Wang F, Yuan J, Wang Z, Li J, Li J, Feng C, Zhang Z, Wang L, Peng L, Chen L, Qin Y, Zhao D, Tan S, Yin L, Xu J, Zhou C, Jiang C, Liu L (2020) Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci China Life Sci 63(3):364–374. https://doi.org/10.1007/s11427-020-1643-8
McElvaney Oliver J, McEvoy Natalie L, McElvaney Oisín F, Carroll Tomás P, Murphy Mark P, Dunlea Danielle M, Choileáin Orna Ní, Clarke Jennifer, O’Connor Eoin, Hogan Grace, Ryan Daniel, Sulaiman Imran, Gunaratnam Cedric, Branagan Peter, O’Brien Michael E, Morgan Ross K, Costello Richard W, Hurley Killian, Walsh Seán, de Barra Eoghan, McNally Cora, McConkey Samuel, Boland Fiona, Galvin Sinead, Kiernan Fiona, O’Rourke James, Dwyer Rory, Power Michael, Geoghegan Pierce, Larkin Caroline, O’Leary Ruth Aoibheann, Freeman James, Gaffney Alan, Marsh Brian, Curley Gerard F, McElvaney NG (2020) Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am J Respir Crit Care Med 202(6):812–821
Ortiz-Brizuela E, Villanueva-Reza M, Gonzalez-Lara MF, Tamez-Torres KM, Roman-Montes CM, Diaz-Mejia BA, Perez-Garcia E, Olivas-Martinez A, Rajme-Lopez S, Martinez-Guerra BA, de-Leon-Cividanes NA, Fernandez-Garcia OA, Guerrero-Torres L, Torres-Gonzalez L, Carrera-Patino FA, Corral-Herrera EA, Hernandez-Alemon AN, Tovar-Vargas MLA, Serrano-Pinto YG, Espejo-Ortiz CE, Morales-Ortega ML, Lozano-Cruz OA, Cardenas-Fragoso JL, Vidal-Mayo JJ, Hernandez-Gilsoul T, Rivero-Sigarroa E, Dominguez-Cherit G, Cervantes-Villar LE, Ramos-Cervantes MDP, Ibarra-Gonzalez V, Calva-Mercado JJ, Sierra-Madero JG, Lopez-Iniguez A, Ochoa-Hein E, Crabtree-Ramirez BE, Galindo-Fraga A, Guerrero-Almeida ML, Ruiz-Palacios GM, Gulias-Herrero A, Sifuentes-Osornio J, Kershenobich-Stalnikowitz D, Ponce-de-Leon A (2020) Clinical and Epidemiological Characteristics of Patients Diagnosed with Covid-19 in a Tertiary Care Center in Mexico City: a Prospective Cohort Study. Rev Invest Clin 72(3):165–177. https://doi.org/10.24875/RIC.20000211
Petrilli CM, Jones SA, Yang J, Rajagopalan H, O’Donnell L, Chernyak Y, Tobin KA, Cerfolio RJ, Francois F, Horwitz LI (2020) Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ 369:m1966. https://doi.org/10.1136/bmj.m1966
Ponziani FR, Del Zompo F, Nesci A, Santopaolo F, Ianiro G, Pompili M, Gasbarrini A, Gemelli Against C-G (2020) Liver involvement is not associated with mortality: results from a large cohort of SARS-CoV-2 positive patients. Aliment Pharmacol Ther. https://doi.org/10.1111/apt.15996
Simone Bastrup Israelsen, Klaus Tjelle Kristiansen, Bettina Hindsberger, Charlotte Suppli Ulrik, Ove Andersen, Magnus Jensen, Steen Andersen, Christian Rasmussen, Henrik L Jørgensen, Christian Østergaard, Bjarne Ørskov Lindhardt, Gitte Kronborg, Benfield T (2020) Characteristics of patients with COVID-19 pneumonia at Hvidovre Hospital, March-April 2020. Danish Med J 67(6)
Suleyman G, Fadel RA, Malette KM, Hammond C, Abdulla H, Entz A, Demertzis Z, Hanna Z, Failla A, Dagher C, Chaudhry Z, Vahia A, Abreu Lanfranco O, Ramesh M, Zervos MJ, Alangaden G, Miller J, Brar I (2020) Clinical characteristics and morbidity associated with Coronavirus Disease 2019 in a series of patients in metropolitan detroit. JAMA Netw Open 3(6):e2012270. https://doi.org/10.1001/jamanetworkopen.2020.12270
Sun DQ, Wang TY, Zheng KI, Targher G, Byrne CD, Chen YP, Zheng MH (2020) Subclinical acute kidney injury in COVID-19 patients: a retrospective cohort study. Nephron 144(7):347–350. https://doi.org/10.1159/000508502
Urra JM, Cabrera CM, Porras L, Rodenas I (2020) Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin Immunol 217:108486. https://doi.org/10.1016/j.clim.2020.108486
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, **ang H, Cheng Z, **ong Y, Zhao Y, Li Y, Wang X, Peng Z (2020) Clinical characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan. JAMA, China. https://doi.org/10.1001/jama.2020.1585
Wang F, Hou H, Luo Y, Tang G, Wu S, Huang M, Liu W, Zhu Y, Lin Q, Mao L, Fang M, Zhang H, Sun Z (2020) The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. https://doi.org/10.1172/jci.insight.137799
Wang R, Pan M, Zhang X, Han M, Fan X, Zhao F, Miao M, Xu J, Guan M, Deng X, Chen X, Shen L (2020) Epidemiological and clinical features of 125 Hospitalized Patients with COVID-19 in Fuyang, Anhui, China. Int J Infect Dis 95:421–428. https://doi.org/10.1016/j.ijid.2020.03.070
Wu J, Huang J, Zhu G, Wang Q, Lv Q, Huang Y, Yu Y, Si X, Yi H, Wang C, Liu Y, **ao H, Zhou Q, Liu X, Yang D, Guan X, Li Y, Peng S, Sung J, **ao H (2020) Elevation of blood glucose level predicts worse outcomes in hospitalized patients with COVID-19: a retrospective cohort study. BMJ Open Diabetes Res Care. https://doi.org/10.1136/bmjdrc-2020-001476
Yang L, Liu J, Zhang R, Li M, Li Z, Zhou X, Hu C, Tian F, Zhou F, Lei Y (2020) Epidemiological and clinical features of 200 hospitalized patients with corona virus disease 2019 outside Wuhan, China: a descriptive study. J Clin Virol 129:104475. https://doi.org/10.1016/j.jcv.2020.104475
Zeng Z, Ma Y, Zeng H, Huang P, Liu W, Jiang M, **ang X, Deng D, Liao X, Chen P, Chen Y (2020) Simple nomogram based on initial laboratory data for predicting the probability of ICU transfer of COVID-19 patients: multicenter retrospective study. J Med Virol. https://doi.org/10.1002/jmv.26244
Zhou Yonggang, Binqing Fu, Zheng **aohu, Wang Dongsheng, Zhao Changcheng, Qi Yingjie, Sun Rui, Tian Zhigang, **aoling Xu, Wei H (2020) Pathogenic T-cells and inflammatory monocytes incite inflammatory storms in severe COVID-19 patients. Natl Sci Rev. https://doi.org/10.1093/nsr/nwaa041
Bao J, Li C, Zhang K, Kang H, Chen W, Gu B (2020) Comparative analysis of laboratory indexes of severe and non-severe patients infected with COVID-19. Clin Chim Acta 509:180–194. https://doi.org/10.1016/j.cca.2020.06.009
Chang TH, Wu JL, Chang LY (2020) Clinical characteristics and diagnostic challenges of pediatric COVID-19: a systematic review and meta-analysis. J Formos Med Assoc 119(5):982–989. https://doi.org/10.1016/j.jfma.2020.04.007
Deng X, Liu B, Li J, Zhang J, Zhao Y, Xu K (2020) Blood biochemical characteristics of patients with coronavirus disease 2019 (COVID-19): a systemic review and meta-analysis. Clin Chem Lab Med 58(8):1172–1181. https://doi.org/10.1515/cclm-2020-0338
Feng X, Li S, Sun Q, Zhu J, Chen B, **ong M, Cao G (2020) Immune-inflammatory parameters in COVID-19 cases: a systematic review and meta-analysis. Front Med (Lausanne) 7:301. https://doi.org/10.3389/fmed.2020.00301
Fu L, Wang B, Yuan T, Chen X, Ao Y, Fitzpatrick T, Li P, Zhou Y, Lin YF, Duan Q, Luo G, Fan S, Lu Y, Feng A, Zhan Y, Liang B, Cai W, Zhang L, Du X, Li L, Shu Y, Zou H (2020) Clinical characteristics of coronavirus disease 2019 (COVID-19) in China: a systematic review and meta-analysis. J Infect 80(6):656–665. https://doi.org/10.1016/j.**f.2020.03.041
Henry BM, Benoit SW, de Oliveira MHS, Hsieh WC, Benoit J, Ballout RA, Plebani M, Lippi G (2020) Laboratory abnormalities in children with mild and severe coronavirus disease 2019 (COVID-19): a pooled analysis and review. Clin Biochem 81:1–8. https://doi.org/10.1016/j.clinbiochem.2020.05.012
Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G (2020) Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chem Lab Med 58(7):1021–1028. https://doi.org/10.1515/cclm-2020-0369
Hozo SP, Djulbegovic B, Hozo I (2005) Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol 5:13. https://doi.org/10.1186/1471-2288-5-13
Huang D, Lian X, Song F, Ma H, Lian Z, Liang Y, Qin T, Chen W, Wang S (2020) Clinical features of severe patients infected with 2019 novel coronavirus: a systematic review and meta-analysis. Ann Transl Med 8(9):576. https://doi.org/10.21037/atm-20-2124
Huang I, Pranata R, Lim MA, Oehadian A, Alisjahbana B (2020) C-reactive protein, procalcitonin, D-dimer, and ferritin in severe coronavirus disease-2019: a meta-analysis. Ther Adv Respir Dis 14:1753466620937175. https://doi.org/10.1177/1753466620937175
Huang X, Wei F, Yang Z, Li M, Liu L, Chen K (2020) Lactose dehydrogenase in patients with severe COVID-19: a meta-analysis of retrospective study. Prehosp Disaster Med 35(4):462–463. https://doi.org/10.1017/S1049023X20000576
Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A (2020) The role of biomarkers in diagnosis of COVID-19 - A systematic review. Life Sci 254:117788. https://doi.org/10.1016/j.lfs.2020.117788
Lima WG, Barra A, Brito JCM, Nizer WSC (2020) D-Dimer serum levels as a biomarker associated for the lethality in patients with coronavirus disease 2019: a meta-analysis. Blood Coagul Fibrinolysis 31(5):335–338. https://doi.org/10.1097/MBC.0000000000000927
Lippi G, Plebani M (2020) Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Clin Chim Acta 505:190–191. https://doi.org/10.1016/j.cca.2020.03.004
Lippi G, Plebani M, Henry BM (2020) Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin Chim Acta 506:145–148. https://doi.org/10.1016/j.cca.2020.03.022
Parohan M, Yaghoubi S, Seraji A (2020) Cardiac injury is associated with severe outcome and death in patients with Coronavirus disease 2019 (COVID-19) infection: a systematic review and meta-analysis of observational studies. Eur Heart J Acute Cardiovasc Care. https://doi.org/10.1177/2048872620937165
Sahu BR, Kampa RK, Padhi A, Panda AK (2020) C-reactive protein: a promising biomarker for poor prognosis in COVID-19 infection. Clin Chim Acta 509:91–94. https://doi.org/10.1016/j.cca.2020.06.013
Shi L, Wang Y, Wang Y, Duan G, Yang H (2020) An updated meta-analysis on the relationship between D-dimer levels and severity of coronavirus disease 2019. Int J Lab Hematol. https://doi.org/10.1111/ijlh.13268
Soraya GV, Ulhaq ZS (2020) Crucial laboratory parameters in COVID-19 diagnosis and prognosis: an updated meta-analysis. Med Clin (Barc) 155(4):143–151. https://doi.org/10.1016/j.medcli.2020.05.017
Taylor EH, Hofmeyr R, Torborg A, van Tonder C, Boden R, Earle E, Nejthardt M, Kabambi KF, Isaacs M, Usenbo A, Gerber C, van der Spuy K, Mrara B, Ndhlovu T, Chen A, Swanevelder J, Coetzee J, Biccard BM (2020) Risk factors and interventions associated with mortality or survival in adult COVID-19 patients admitted to critical care: a systematic review and meta-analysis. South Afr J Anaesth Analg. https://doi.org/10.36303/sajaa.2020.26.3.2428
Tian W, Jiang W, Yao J, Nicholson CJ, Li RH, Sigurslid HH, Wooster L, Rotter JI, Guo X, Malhotra R (2020) Predictors of mortality in hospitalized COVID-19 patients: a systematic review and meta-analysis. J Med Virol. https://doi.org/10.1002/jmv.26050
Toraih EA, Elshazli RM, Hussein MH, Elgaml A, Amin M, El-Mowafy M, El-Mesery M, Ellythy A, Duchesne J, Killackey MT, Ferdinand KC, Kandil E, Fawzy MS (2020) Association of cardiac biomarkers and comorbidities with increased mortality, severity, and cardiac injury in COVID-19 patients: a meta-regression and decision tree analysis. J Med Virol. https://doi.org/10.1002/jmv.26166
**ong M, Liang X, Wei YD (2020) Changes in blood coagulation in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. Br J Haematol 189(6):1050–1052. https://doi.org/10.1111/bjh.16725
Yamada T, Wakabayashi M, Yamaji T, Chopra N, Mikami T, Miyashita H, Miyashita S (2020) Value of leukocytosis and elevated C-reactive protein in predicting severe coronavirus 2019 (COVID-19): a systematic review and meta-analysis. Clin Chim Acta 509:235–243. https://doi.org/10.1016/j.cca.2020.06.008
Zeng F, Huang Y, Guo Y, Yin M, Chen X, **ao L, Deng G (2020) Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int J Infect Dis 96:467–474. https://doi.org/10.1016/j.ijid.2020.05.055
Zhang H, Wu T (2020) CD4 + T, CD8 + T counts and severe COVID-19: a meta-analysis. J Infect 81(3):e82–e84. https://doi.org/10.1016/j.**f.2020.06.036
Zhang ZL, Hou YL, Li DT, Li FZ (2020) Laboratory findings of COVID-19: a systematic review and meta-analysis. Scand J Clin Lab Invest. https://doi.org/10.1080/00365513.2020.1768587
Zheng Z, Peng F, Xu B, Zhao J, Liu H, Peng J, Li Q, Jiang C, Zhou Y, Liu S, Ye C, Zhang P, **ng Y, Guo H, Tang W (2020) Risk factors of critical & mortal COVID-19 cases: a systematic literature review and meta-analysis. J Infect 81(2):e16–e25. https://doi.org/10.1016/j.**f.2020.04.021
Zhu J, Pang J, Ji P, Zhong Z, Li H, Li B, Zhang J (2020) Elevated interleukin-6 is associated with severity of COVID-19: a meta-analysis. J Med Virol. https://doi.org/10.1002/jmv.26085
Channappanavar R, Zhao J, Perlman S (2014) T cell-mediated immune response to respiratory coronaviruses. Immunol Res 59(1–3):118–128. https://doi.org/10.1007/s12026-014-8534-z
Shaw AC, Goldstein DR, Montgomery RR (2013) Age-dependent dysregulation of innate immunity. Nat Rev Immunol 13(12):875–887. https://doi.org/10.1038/nri3547
Liu WJ, Zhao M, Liu K, Xu K, Wong G, Tan W, Gao GF (2017) T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antiviral Res 137:82–92. https://doi.org/10.1016/j.antiviral.2016.11.006
Lr FA (2020) Neutrophil-to-lymphocyte ratio and lymphocyte-to-C-reactive protein ratio in patients with severe coronavirus disease 2019 (COVID-19): a meta-analysis. J Med Virol 170:45–52. https://doi.org/10.1016/j.thromres.2018.08.002
Wu C, Chen X, Cai Y, **a J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, **ong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y (2020) Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan. JAMA Intern Med, China. https://doi.org/10.1001/jamainternmed.2020.0994
Mehta P, McAuley DF, Brown M, Sanchez E, Tattersall RS, Manson JJ (2020) COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet 395(10229):1033–1034. https://doi.org/10.1016/s0140-6736(20)30628-0
Feld J, Tremblay D, Thibaud S, Kessler A, Naymagon L (2020) Ferritin levels in patients with COVID-19: a poor predictor of mortality and hemophagocytic lymphohistiocytosis. Int J Lab Hematol. https://doi.org/10.1111/ijlh.13309
Henter JI, Horne A, Arico M, Egeler RM, Filipovich AH, Imashuku S, Ladisch S, McClain K, Webb D, Winiarski J, Janka G (2007) HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 48(2):124–131. https://doi.org/10.1002/pbc.21039
Wood H, Jones JR, Hui K, Mare T, Pirani T, Galloway J, Metaxa V, Benjamin R, Rutherford A, Cain S, Kulasekararaj AG (2020) Secondary HLH is uncommon in severe COVID-19. Br J Haematol. https://doi.org/10.1111/bjh.16934
Mahallawi WH, Khabour OF, Zhang Q, Makhdoum HM, Suliman BA (2018) MERS-CoV infection in humans is associated with a pro-inflammatory Th1 and Th17 cytokine profile. Cytokine 104:8–13. https://doi.org/10.1016/j.cyto.2018.01.025
Sager R, Kutz A, Mueller B, Schuetz P (2017) Procalcitonin-guided diagnosis and antibiotic stewardship revisited. BMC Med 15(1):15. https://doi.org/10.1186/s12916-017-0795-7
Yang MLC, Li K, Ho KML, Ng MHL, Chan KSC, Fok TF (2004) Hematological findings in SARS patients and possible mechanisms (Review). Int J Mol Med 14(2):311–315
Fan BE, Chong VCL, Chan SSW, Lim GH, Lim KGE, Tan GB, Mucheli SS, Kuperan P, Ong KH (2020) Hematologic parameters in patients with COVID-19 infection. Am J Hematol. https://doi.org/10.1002/ajh.25774
Yang X, Yang Q, Wang Y, Wu Y, Xu J, Yu Y, Shang Y (2020) Thrombocytopenia and its association with mortality in patients with COVID-19. J Thromb Haemost. https://doi.org/10.1111/jth.14848
Levi M, Thachil J, Iba T, Levy JH (2020) Coagulation abnormalities and thrombosis in patients with COVID-19. The Lancet Haematol 7(6):e438–e440. https://doi.org/10.1016/s2352-3026(20)30145-9
Naymagon L, Zubizarreta N, Feld J, van Gerwen M, Alsen M, Thibaud S, Kessler A, Venugopal S, Makki I, Qin Q, Dharmapuri S, Jun T, Bhalla S, Berwick S, Christian K, Mascarenhas J, Dembitzer F, Moshier E, Tremblay D (2020) Admission D-dimer levels, D-dimer trends, and outcomes in COVID-19. Thromb Res 196:99–105. https://doi.org/10.1016/j.thromres.2020.08.032
Acknowledgements
This work was funded by the Human Resources Development Operational Programme Grant, Grant Number: EFOP–3.6.2–16–2017–00006 – LIVE LONGER is co-financed by the European Union (European Regional Development Fund) within the framework of Programme Széchenyi 2020.
Funding
Open access funding provided by University of Pécs.. This work was funded by the Human Resources Development Operational Programme Grant, Grant Number: EFOP–3.6.2–16–2017–00006 – LIVE LONGER is co-financed by the European Union (European Regional Development Fund) within the framework of Programme Széchenyi 2020. The funder of the study had no role in study design, data collection, data analysis, data interpretation, or the writing of the report.
Author information
Authors and Affiliations
Contributions
SK: preparation of the draft of the manuscript, selection of studies, data extraction, risk of bias assessment; GN: statistical analysis, preparation of the standardized data collection sheet; PH: substantial contribution in study design; DN: statistical analysis; MF: selection of studies, data extraction; FD: selection of studies, data extraction; BN: data extraction, risk of bias assessment; MFJ: selection of studies, preparation of the standardized data collection sheet, stylistic and grammatical revision of the manuscript; KO: risk of bias assessment, stylistic and grammatical revision of the manuscript; ZM: expert in the field of anaesthesiology and intensive therapy, substantial contribution in study design and interpretation of data, preparation of the manuscript; NZ: substantial contribution in study design, validation of the data extraction; AP: preparation of the study protocol; PJH: preparation of the standardized data collection sheet, stylistic and grammatical revision of the manuscript; ZS: participation in the design of the study and its coordination; PG: provided revisions to the scientific content of the manuscript; BE: provided revisions to the scientific content of the manuscript; AH: expert in the field of haematology, substantial contribution in study design and interpretation of data, preparation of study protocol and the first draft of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors do not have any conflicts of interest to declare.
Ethical approval
This study was prepared in accordance with the Committee on Publication Ethics (COPE) guidelines to respect third parties rights such as copyright and/or moral rights. Ethical approval was not required to conduct this project as data is not individualized and primary data was not collected. Not required as data is not individualized and primary data was not collected.
Consent to participate
Not required as data is not individualized and primary data was not collected.
Consent to publish
The corresponding author accepts responsibility for releasing this material on behalf of any and all co-authors.
Additional information
Edited by Matthias J. Reddehase.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kiss, S., Gede, N., Hegyi, P. et al. Early changes in laboratory parameters are predictors of mortality and ICU admission in patients with COVID-19: a systematic review and meta-analysis. Med Microbiol Immunol 210, 33–47 (2021). https://doi.org/10.1007/s00430-020-00696-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00430-020-00696-w