Abstract
Objectives
To assess the value of coordinatized lesion location analysis (CLLA), in empowering ROI-based imaging diagnosis of gliomas by improving accuracy and generalization performances.
Methods
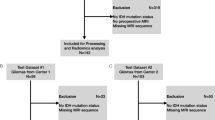
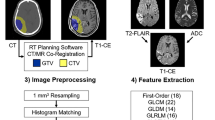
In this retrospective study, pre-operative contrasted T1-weighted and T2-weighted MR images were obtained from patients with gliomas from three centers: **ling Hospital, Tiantan Hospital, and the Cancer Genome Atlas Program. Based on CLLA and ROI-based radiomic analyses, a fusion location-radiomics model was constructed to predict tumor grades, isocitrate dehydrogenase (IDH) status, and overall survival (OS). An inter-site cross-validation strategy was used for assessing the performances of the fusion model on accuracy and generalization with the value of area under the curve (AUC) and delta accuracy (ACC) (ACCtesting—ACCtraining). Comparisons of diagnostic performances were performed between the fusion model and the other two models constructed with location and radiomics analysis using DeLong’s test and Wilcoxon signed ranks test.
Results
A total of 679 patients (mean age, 50 years ± 14 [standard deviation]; 388 men) were enrolled. Based on tumor location probabilistic maps, fusion location-radiomics models (averaged AUC values of grade/IDH/OS: 0.756/0.748/0.768) showed the highest accuracy in contrast to radiomics models (0.731/0.686/0.716) and location models (0.706/0.712/0.740). Notably, fusion models ([median Delta ACC: − 0.125, interquartile range: 0.130]) demonstrated improved generalization than that of radiomics model ([− 0.200, 0.195], p = 0.018).
Conclusions
CLLA could empower ROI-based radiomics diagnosis of gliomas by improving the accuracy and generalization of the models.
Clinical relevance statement
This study proposed a coordinatized lesion location analysis for glioma diagnosis, which could improve the performances of the conventional ROI-based radiomics model in accuracy and generalization.
Key Points
• Using coordinatized lesion location analysis, we mapped anatomic distribution patterns of gliomas with specific pathological and clinical features and constructed glioma prediction models.
• We integrated coordinatized lesion location analysis into ROI-based analysis of radiomics to propose new fusion location-radiomics models.
• Fusion location-radiomics models, with the advantages of being less influenced by variabilities, improved accuracy, and generalization performances of ROI-based radiomics models on predicting the diagnosis of gliomas.






Similar content being viewed by others
Abbreviations
- AUC:
-
Area under the curve
- HGG:
-
High-grade glioma
- IDH:
-
Isocitrate dehydrogenase
- LGG:
-
Low-grade glioma
- OS:
-
Overall survival
- ROI:
-
Region-of-interest
- TCGA:
-
The Cancer Genome Atlas
- WHO:
-
World Health Organization
References
Low JT, Ostrom QT, Cioffi G et al (2022) Primary brain and other central nervous system tumors in the United States (2014–2018): a summary of the CBTRUS statistical report for clinicians. Neurooncol Pract 9:165–182
Germann J, Zadeh G, Mansouri A, Kucharczyk W, Lozano AM, Boutet A (2022) Untapped Neuroimaging Tools for Neuro-Oncology: Connectomics and Spatial Transcriptomics. Cancers 14:464
Hu LS, Hawkins-Daarud A, Wang LJ, Li J, Swanson KR (2020) Imaging of intratumoral heterogeneity in high-grade glioma. Cancer Lett 477:97–106
Boonzaier NR, Larkin TJ, Matys T, van der Hoorn A, Yan JL, Price SJ (2017) Multiparametric MR imaging of diffusion and perfusion in contrast-enhancing and nonenhancing components in patients with glioblastoma. Radiology 284:180–190
Bette S, Huber T, Gempt J et al (2017) Local fractional anisotropy is reduced in areas with tumor recurrence in glioblastoma. Radiology 283:499–507
Karaman MM, Zhang JX, **e KL, Zhu WZ, Zhou XHJ (2021) Quartile histogram assessment of glioma malignancy using high b-value diffusion MRI with a continuous-time random-walk model. NMR Biomed 34:e4485
Zhang S, Chiang GCY, Magge RS et al (2019) Texture analysis on conventional MRI images accurately predicts early malignant transformation of low-grade gliomas. Eur Radiol 29:2751–2759
Tomaszewski MR, Gillies RJ (2021) The biological meaning of radiomic features. Radiology 298:505–516
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
Le NQK, Hung TNK, Do DT, Lam LHT, Dang LH, Huynh TT (2021) Radiomics-based machine learning model for efficiently classifying transcriptome subtypes in glioblastoma patients from MRI. Comput Biol Med 132:104320
Sudre CH, Panovska-Griffiths J, Sanverdi E et al (2020) Machine learning assisted DSC-MRI radiomics as a tool for glioma classification by grade and mutation status. BMC Med Inform Decis Mak 20:149
Malik N, Geraghty B, Dasgupta A et al (2021) MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J Neurooncol 155:181–191
Li YM, Wei D, Liu X et al (2022) Molecular subty** of diffuse gliomas using magnetic resonance imaging: comparison and correlation between radiomics and deep learning. Eur Radiol 32:747–758
Kim M, Jung SY, Park JE et al (2020) Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur Radiol 30:2142–2151
Choi YS, Bae S, Chang JH et al (2021) Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol 23:304–313
Zhuo ZZ, Qu LY, Zhang P et al (2021) Prediction of H3K27M-mutant brainstem glioma by amide proton transfer-weighted imaging and its derived radiomics. Eur J Nucl Med Mol Imaging 48:4426–4436
Lam LHT, Do DT, Diep DTN et al (2022) Molecular subtype classification of low-grade gliomas using magnetic resonance imaging-based radiomics and machine learning. NMR Biomed 35:e4792
Yan J, Zhaoc YS, Chend YS et al (2021) Deep learning features from diffusion tensor imaging improve glioma stratification and identify risk groups with distinct molecular pathway activities. EBioMedicine 72:103583
Vajapeyam S, Brown D, Billups C et al (2020) Advanced ADC histogram, perfusion, and permeability metrics show an association with survival and pseudoprogression in newly diagnosed diffuse intrinsic pontine glioma: a report from the pediatric brain tumor consortium. AJNR Am J Neuroradiol 41:718–724
Vecchio TG, Neimantaite A, Corell A et al (2021) Lower-grade gliomas: an epidemiological voxel-based analysis of location and proximity to eloquent regions. Front Oncol 11:748229
Fyllingen EH, Bo LE, Reinertsen I et al (2021) Survival of glioblastoma in relation to tumor location: a statistical tumor atlas of a population-based cohort. Acta Neurochir 163:1895–1905
Steed TC, Treiber JM, Taha B et al (2020) Glioblastomas located in proximity to the subventricular zone (SVZ) exhibited enrichment of gene expression profiles associated with the cancer stem cell state. J Neurooncol 148:455–462
Cardinale F, Cossu M, Castana L et al (2013) Stereoelectroencephalography: surgical methodology, safety, and stereotactic application accuracy in 500 procedures. Neurosurgery 72:353–366
Fan X, Wang YY, Liu Y et al (2016) Brain regions associated with telomerase reverse transcriptase promoter mutations in primary glioblastomas. J Neurooncol 128:455–462
Liu TT, Achrol AS, Mitchell LA et al (2016) Computational identification of tumor anatomic location associated with survival in 2 large cohorts of human primary glioblastomas. AJNR Am J Neuroradiol 37:621–628
Neyra MAT, Neuberger U, Reinhardt A et al (2018) Voxel-wise radiogenomic map** of tumor location with key molecular alterations in patients with glioma. Neuro Oncol 20:1517–1524
Tang QS, Lian YX, Yu JH, Wang YY, Shi ZF, Chen L (2017) Anatomic map** of molecular subtypes in diffuse glioma. BMC Neurol 17:183
Yunhe M, Yuan Y, ** seizure foci and tumor genetic factors in glioma associated seizure patients. J Neurosurg Sci 64:456–463
Ellingson BM, Lai A, Harris RJ et al (2013) Probabilistic radiographic atlas of glioblastoma phenotypes. AJNR Am J Neuroradiol 34:533–540
Zwanenburg A, Vallieres M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenoty**. Radiology 295:328–338
Meyer M, Ronald J, Vernuccio F et al (2019) Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology 293:583–591
Berenguer R, Pastor-Juan MD, Canales-Vazquez J et al (2018) Radiomics of CT features may be nonreproducible and redundant: influence of CT acquisition parameters. Radiology 288:407–415
Mazziotta J, Toga A, Evans A et al (2001) A probabilistic atlas and reference system for the human brain: International Consortium for Brain Map** (ICBM). Philos Trans R Soc Lond B Biol Sci 356:1293–1322
Brett M, Leff AP, Rorden C, Ashburner J (2001) Spatial normalization of brain images with focal lesions using cost function masking. Neuroimage 14:486–500
Wu D, Ma T, Ceritoglu C et al (2016) Resource atlases for multi-atlas brain segmentations with multiple ontology levels based on T1-weighted MRI. Neuroimage 125:120–130
Segato A, Marzullo A, Calimeri F, De Momi E (2020) Artificial intelligence for brain diseases: a systematic review. APL Bioeng 4:041503
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Hu XF, Gong J, Zhou W et al (2021) Computer-aided diagnosis of ground glass pulmonary nodule by fusing deep learning and radiomics features. Phys Med Biol 66:065015
Li A, Zalesky A, Yue WH et al (2020) A neuroimaging biomarker for striatal dysfunction in schizophrenia. Nat Med 26:558–565
Delong ER, Delong DM, Clarkepearson DI (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Müller DMJ, Robe PA, Ardon H et al (2020) Quantifying eloquent locations for glioblastoma surgery using resection probability maps. J Neurosurg 134:1091–1101
Roux A, Roca P, Edjlali M et al (2019) MRI Atlas of IDH wild-type supratentorial glioblastoma: probabilistic maps of phenotype, management, and outcomes. Radiology 293:633–643
Lim DA, Cha S, Mayo MC et al (2007) Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol 9:424–429
Park CJ, Han K, Kim H et al (2020) Radiomics risk score may be a potential imaging biomarker for predicting survival in isocitrate dehydrogenase wild-type lower-grade gliomas. Eur Radiol 30:6464–6474
Nakamoto T, Takahashi W, Haga A et al (2020) Prediction of malignant glioma grades using contrast-enhanced T1-weighted and T2-weighted magnetic resonance images based on a radiomic analysis. Sci Rep 9:19411
Acknowledgements
Thanks are due to John W Chen from the Massachusetts General Hospital, Harvard Medical School for the revision of our manuscript.
Funding
This study has received funding by grants from the National Key Technology (R&D) Program of the Ministry of Science and Technology (2018YFA0701703 to Zhiqiang Zhang); the National Science and Technology Innovation 2030 − Major program of “Brain Science and Brain-Like Research” (2022ZD0211800 to Zhiqiang Zhang); Xuzhou Medical University Open Fund Project (XYKF202101 to Zhiqiang Zhang); the National Key R&D Program of China (2020AAA0109505 to Guangming Lu); and the National Natural Science Foundation of China (82127806 to Guangming Lu).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Zhiqiang Zhang from the Department of Diagnostic Radiology, Affiliated **ling Hospital, Medical School of Nan**g University.
Conflict of interest
Qing Zhou and Feng Shi are employees of Shanghai United Imaging Intelligence Co., Ltd. The company has no role in designing and performing the surveillance and analyzing and interpreting the data. The other authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
Mengjie Lu from the School of Public Health, Shanghai Jiaotong University School of Medicine, is one of the authors who has significant statistical expertise.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• retrospective
• cross-sectional study
• multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Zhang, Q., Li, J. et al. Coordinatized lesion location analysis empowering ROI-based radiomics diagnosis on brain gliomas. Eur Radiol 33, 8776–8787 (2023). https://doi.org/10.1007/s00330-023-09871-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09871-y