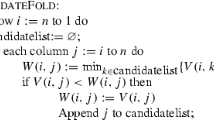Abstract
RNA secondary structure folding kinetics is known to be important for the biological function of certain processes, such as the hok/sok system in E. coli. Although linear algebra provides an exact computational solution of secondary structure folding kinetics with respect to the Turner energy model for tiny (\(\approx \)20 nt) RNA sequences, the folding kinetics for larger sequences can only be approximated by binning structures into macrostates in a coarse-grained model, or by repeatedly simulating secondary structure folding with either the Monte Carlo algorithm or the Gillespie algorithm. Here we investigate the relation between the Monte Carlo algorithm and the Gillespie algorithm. We prove that asymptotically, the expected time for a K-step trajectory of the Monte Carlo algorithm is equal to \(\langle N \rangle \) times that of the Gillespie algorithm, where \(\langle N \rangle \) denotes the Boltzmann expected network degree. If the network is regular (i.e. every node has the same degree), then the mean first passage time (MFPT) computed by the Monte Carlo algorithm is equal to MFPT computed by the Gillespie algorithm multiplied by \(\langle N \rangle \); however, this is not true for non-regular networks. In particular, RNA secondary structure folding kinetics, as computed by the Monte Carlo algorithm, is not equal to the folding kinetics, as computed by the Gillespie algorithm, although the mean first passage times are roughly correlated. Simulation software for RNA secondary structure folding according to the Monte Carlo and Gillespie algorithms is publicly available, as is our software to compute the expected degree of the network of secondary structures of a given RNA sequence—see http://bioinformatics.bc.edu/clote/RNAexpNumNbors.










Similar content being viewed by others
Notes
Definitions of Monte Carlo algorithm, Gillespie algorithm, detailed balance, mean first passage time, master equation, etc. are given later in the paper.
Each step of Algorithms 2, 3 and 4 involves a transition from a current state to a distinct state; however, each step of the time-driven Monte Carlo Algorithm 1 does not necessarily involve a transition to a new state, especially if the current state x has low energy, so that \(p_{x,x}\) may be large. For this reason, we use the term K-transition trajectory.
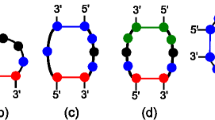
Recall that secondary structures may be represented as a set of base pairs.
ME stands for the Master Equation, meaning the computation of the time-dependent population occupancy vector \(\mathbf{p}(t)\) by solution of the master equation \(\frac{d \mathbf{p}(t)}{dt} = \mathbf{p}(t) \cdot Q\), where Q is the rate matrix. MC stands for Monte Carlo simulation, and MMC stands for Map-based Monte Carlo simulation, a method inspired by probabilistic road map methods from robotics, where a constant k many closest neighboring secondary structures are selected for each RNA secondary structure from a sampled collection of modest size. Subsequently Monte Carlo simulation is applied to the resulting network, which is much smaller than the network of all secondary structures. The correctness of the authors’ assertion is due uniquely to the fact that the roadmap sampling network is k-regular.
References
Abkevich VI, Gutin AM, Shakhnovich EI (1996) How the first biopolymers could have evolved. Proc Natl Acad Sci USA 93(2):839–844
Abkevich VI, Gutin AM, Shakhnovich EI (1997) Computer simulations of prebiotic evolution. Pac Symp Biocomput 27–38
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230
Aviram I, Veltman I, Churkin A, Barash D (2012) Efficient procedures for the numerical simulation of mid-size RNA kinetics. Algorithms Mol Biol 7(1):24
Bowman GR (1084) A tutorial on building markov state models with MSMBuilder and coarse-graining them with BACE. Methods Mol Biol 141–158:2014
Bowman GR, Huang X, Pande VS (2009) Using generalized ensemble simulations and Markov state models to identify conformational states. Methods 49(2):197–201
Bowman GR, Pande VS (2010) Protein folded states are kinetic hubs. Proc Natl Acad Sci USA 107(24):10890–10895
Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG (1995) Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins 21(3):167–195
Choi HM, Beck VA, Pierce NA (2014) Multiplexed in situ hybridization using hybridization chain reaction. Zebrafish 11(5):488–489
Clote P (2015) Expected degree for RNA secondary structure networks. J Comput Chem 36(2):103–17
Clote P, Backofen R (2000) Computational molecular biology: an introduction. Wiley, Hoboken
Clote P, Bayegan A (2015) Network properties of the ensemble of RNA structures. PLoS ONE 10(10):e0139476
Dotu I, Garcia-Martin JA, Slinger BL, Mechery V, Meyer MM, Clote P (2015) Complete RNA inverse folding: computational design of functional hammerhead ribozymes. Nucleic Acids Res 42(18):11752–11762
Dykeman EC (2015) An implementation of the Gillespie algorithm for RNA kinetics with logarithmic time update. Nucleic Acids Res 43(12):5708–5715
Esmaili-Taheri A, Ganjtabesh M (2015) ERD: a fast and reliable tool for RNA design including constraints. BMC Bioinform 16(1):20
Feller W (1968) An introduction to probability theory and its applications, vol 1, 3rd edn. Wiley, New York
Flamm C, Fontana W, Hofacker IL, Schuster P (2000) RNA folding at elementary step resolution. RNA 6:325–338
Fusy E, Clote P (2012) Combinatorics of locally optimal RNA secondary structures. J Math Biol 68(1–2):341–75
Garcia-Martin JA, Clote P, Dotu I (2013) RNAiFold: A constraint programming algorithm for RNA inverse folding and molecular design. J Bioinform Comput Biol 11(2):1350001. doi:10.1142/S0219720013500017
Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, Bateman A (2011) Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res 39(Database):D141–D145
Geis M, Flamm C, Wolfinger MT, Tanzer A, Hofacker IL, Middendorf M, Mandl C, Stadler PF, Thurner C (2008) Folding kinetics of large RNAs. J Mol Biol 379(1):160–173
Gillespie DT (1976) A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J Comput Phys 22(403):403–434
Harrigan MP, Sultan MM, Hernandez CX, Husic BE, Eastman P, Schwantes CR, Beauchamp KA, McGibbon RT, Pande VS (2017) MSMBuilder: statistical models for biomolecular dynamics. Biophys J 112(1):10–15
Hastings WK (1970) Monte carlo sampling methods using markov chains and their applications. Biometrika 57(1):97–109
Huang X, Yao Y, Bowman GR, Sun J, Guibas LJ, Carlsson G, Pande VS (2010) Constructing multi-resolution Markov State Models (MSMs) to elucidate RNA hairpin folding mechanisms. Pac Symp Biocomput 228–239
Huynen M, Gutell R, Konings D (1997) Assessing the reliability of RNA folding using statistical mechanics. J Mol Biol 267(5):1104–1112
Tinoco I Jr, Schmitz M (2000) Thermodynamics of formation of secondary structure in nucleic acids. In: Di Cera E (ed) Thermodynamics in biology. Oxford University Press, Oxford, pp 131–176
Isambert H, Siggia ED (2000) Modeling RNA folding paths with pseudoknots: application to hepatitis \(delta\) virus ribozyme. Proc Natl Acad Sci USA 97(12):6515–6520
Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37(Database):D159–D162
Klemm K, Flamm C, Stadler PF (2008) Funnels in energy landscapes. Eur Phys J B 63(3):387–391
Levinthal C (1968) Are there pathways for protein folding? Journal de Chimie Physique 65:44–45
Levy RM, Dai W, Deng NJ, Makarov DE (2013) How long does it take to equilibrate the unfolded state of a protein? Protein Sci 22(11):1459–1465
Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) Viennarna package 2.0. Algorithms Mol Biol 6:26
Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21:1087–1092
Meyer CD (1975) The role of the group inverse in the theory of finite Markov chains. SIAM Rev 17(46):443–464
Moulton V, Zuker M, Steel M, Pointon R, Penny D (2000) Metrics on RNA secondary structures. J Comput Biol 7:277–292
Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD (2015) Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43(Database):D130–D137
Nussinov R, Jacobson AB (1980) Fast algorithm for predicting the secondary structure of single stranded RNA. Proc Natl Acad Sci USA 77(11):6309–6313
Pande VS, Beauchamp K, Bowman GR (2010) Everything you wanted to know about Markov State models but were afraid to ask. Methods 52(1):99–105
Šali A, Shakhnovich E, Karplus M (1994a) How does a protein fold? Nature 369(6477):248–251
Šali A, Shakhnovich E, Karplus M (1994b) Kinetics of protein folding: a lattice model study of the requirements for folding to the native state. J Mol Biol 235:1614–1636
Schmitz M, Steger G (1996) Description of RNA folding by “simulated annealing”. J Mol Biol 255(1):254–266
Senter E, Clote P (2015) Fast, approximate kinetics of RNA folding. J Comput Biol 22(2):124–144
Senter E, Sheikh S, Dotu I, Ponty Y, Clote P (2012) Using the fast Fourier transform to accelerate the computational search for RNA conformational switches. PLoS ONE 7(12):e50506
Senter E, Dotu I, Clote P (2015) RNA folding pathways and kinetics using 2D energy landscapes. J Math Biol 70(1–2):173–196
Stein PR, Waterman MS (1978) On some new sequences generalizing the Catalan and Motzkin numbers. Discrete Math 26:261–272
Swope WC, Pitera JW, Suits F (2010) Describing protein folding kinetics by molecular dynamics simulations. 1. Theory. J Phys Chem B 108(21):6571–6581 :228-39
Taneda A (2011) MODENA: a multi-objective RNA inverse folding. Adv Appl Bioinform Chem 4:1–12
Tang X, Thomas S, Tapia L, Amato NM (2007) Tools for simulating and analyzing RNA folding kinetics.In: Speed T, Huang H (eds) Research in computational molecular biology. RECOMB 2007. Lecture notes in computer science, vol 4453. Springer, Berlin, Heidelberg, pp 268–282
Tang X, Thomas S, Tapia L, Giedroc DP, Amato NM (2008) Simulating RNA folding kinetics on approximated energy landscapes. J Mol Biol 381(4):1055–1067
Turner DH, Mathews DH (2010) NNDB: the nearest neighbor parameter database for predicting stability of nucleic acid secondary structure. Nucleic Acids Res 38(Database):D280–D282
Weber JK, Pande VS (2011) Characterization and rapid sampling of protein folding Markov state model topologies. J Chem Theory Comput 7(10):3405–3411
Wolfinger M, Svrcek-Seiler WA, Flamm C, Stadler PF (2004a) Efficient computation of RNA folding dynamics. J Phys A Math Gen 37:4731–4741
Wolfinger MT, Svrcek-Seiler WA, Flamm C, Hofacker IL, Stadler PF (2004b) Efficient folding dynamics of RNA secondary structures. J Phys A Math Gen 37:4731–4741
Wuchty S, Fontana W, Hofacker IL, Schuster P (1999) Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers 49:145–165
Xu X, Yu T, Chen SJ (2016) Understanding the kinetic mechanism of RNA single base pair formation. Proc Natl Acad Sci USA 113(1):116–121
Zadeh JN, Wolfe BR, Pierce NA (2011) Nucleic acid sequence design via efficient ensemble defect optimization. J Comput Chem 32(3):439–452
Zhang W, Chen SJ (2002) RNA hairpin-folding kinetics. Proc Natl Acad Sci USA 99(4):1931–1936
Zuker M, Stiegler P (1981) Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9:133–148
Zuker M, Sankoff D (1984) RNA secondary structures and their prediction. Bull Biol 46(4):591–621
Acknowledgements
We would like to thank anonymous referees for helpful remarks. This research was begun during a visit to California Institute of Technology, Free University of Berlin and the Max Planck Institute for Molecular Genetics. For discussions, P.C. would like to thank Frank Noé, Knut Reinert, Martin Vingron and Marcus Weber (Berlin) and Niles Pierce, Erik Winfree (Pasadena). The research was funded by a Guggenheim Fellowship, Deutscher Akademischer Austauschdienst (DAAD), and by National Science Foundation Grant DBI-1262439. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Research supported by National Science Foundation Grant DBI-1262439.
Appendix
Appendix
1.1 A Markov chain of RNA secondary structures and detailed balance
In this section, we prove that detailed balance does not hold for the Markov chain \(\mathcal {M}_1\) of secondary structures for an RNA sequence, where transition probabilities are defined by Eq. (2). The failure of detailed balance is because for almost any RNA sequence, there are secondary structures x, y that are neighbors of each other, but number N(x) of neighbors of x does not equal the number N(y) of neighbors of y. If every secondary structure of a given RNA sequence has the same number of neighbors, then it is easy to show that the Boltzmann distribution \((\pi _1,\ldots ,\pi _n)\) is the stationary distribution, and a small calculation then shows that detailed balance holds. If different structures have different numbers of neighbors, then the Boltzmann distribution is not in general the stationary distribution. To produce an example where detailed balance fails, we must compute the stationary distribution.
For convenience in following the computation, we first present a simple example using the Nussinov energy model, in which the energy E(s) of a structure is defined to be \(-1\) multiplied by the number of base pairs. Historically, the Nussinov algorithm and energy model (Nussinov and Jacobson 1980) predate the Zuker algorithm (Zuker and Stiegler 1981) and Turner energy model (Turner and Mathews 2010).
Example 3
(Detailed balance does not hold for Nussinov energy model) Consider the 6-nt RNA sequence GGGCCC of length 6, which has only four secondary structures, each having N(s) neighbors, given in dot-bracket notation as follows:
-
1.
\(\bullet \bullet \bullet \bullet \bullet \bullet \): the empty structure \(s_1\) with no base pairs and \(N(s_1)=3\) neighbors.
-
2.
\(\,{\texttt {(}}\,\bullet \bullet \bullet \bullet \,{\texttt {)}}\,\): the structure \(s_2\) with base pair (1, 6) and \(N(s_2)=1\) neighbor.
-
3.
\(\,{\texttt {(}}\,\bullet \bullet \bullet \,{\texttt {)}}\,\bullet \): the structure \(s_3\) with base pair (1, 5) and \(N(s_3)=1\) neighbor.
-
4.
\(\bullet \,{\texttt {(}}\,\bullet \bullet \bullet \,{\texttt {)}}\,\): the structure \(s_4\) with base pair (2, 6) and \(N(s_4)=1\) neighbor.
The transition probability matrix M is given as follows, where we compute the values to 20 decimal places using Eq. (2), but display only 4 decimal places for notational convenience:
Let \(M^{\infty } = \lim \nolimits _{n \rightarrow \infty } M^n\). The stationary probability distribution for M must satisfy \(M (p_1^*, p_2^*, p_3^*, p_4^*)^T = (p_1^*, p_2^*, p_3^*, p_4^*)^T\), where T denotes transpose, and so
Using the actual 20-place values of the transition probability matrix M, whose 4-place approximations are given in Eq. (21), we obtain the millionth power of \(M^{(10^6)}\) by a Mathematica computation. The computed 16-place values are shown to 4-place accuracy in the following:
so that the 4-place approximations of the stationary probabilities are as follows: \(p_1^* = 0.1648\), \(p_2^* = 0.2784\), \(p_3^* = 0.2784\), \(p_4^* = 0.2784\). We then compute that \(p_1^* \cdot p_{1,2} = 0.0550\), while \(p_2^* \cdot p_{2,1} = 0.0617\), which shows that detailed balance does not hold.
Using the Turner energy model (Turner and Mathews 2010), the same example yields a different transition probability matrix (not shown), whose millionth power is as follows:
We find that \(p_1^* =0.9994655553038468164 \approx 0.9995\), \(p_2^* =0.000430315331989701045645180244 \approx 0.0004\), \(p_{1,2}=0.001288878388504780024906293256 \approx 0.0013\), \(p_{2,1}=0.061737166286699882156163710079 \approx 0.617\), and that \(p_1^* \cdot p_{1,2} = 0.0012881895542860573 \approx 0.00129\) while \(p_2^* \cdot p_{2,1} = 0.00002656644920676464 \approx 0.00003\). It follows that detailed balance does not hold for the Turner energy model. For RNA sequences of length n, there are exponentially many (\(\approx \)1.95947 \( \cdot n^{-3/2} \cdot 1.86603^n\)) secondary structures, so similar computations cannot be carried out, and it may be the case that detailed balance “approximately” holds.
Rights and permissions
About this article
Cite this article
Clote, P., Bayegan, A.H. RNA folding kinetics using Monte Carlo and Gillespie algorithms. J. Math. Biol. 76, 1195–1227 (2018). https://doi.org/10.1007/s00285-017-1169-7
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00285-017-1169-7
Keywords
- RNA secondary structure
- Mean first passage time
- Refolding kinetics
- Metropolis algorithm
- Gillespie algorithm mean recurrence time
- Expected network degree