Abstract
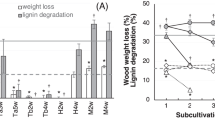
White-rot fungi are the main decomposers of wood cell-wall polymer in forest ecosystems. Little is known, however, about the interactions between white-rot fungi and other coexisting microorganisms in decayed wood. A white-rot fungus, Trametes versicolor strain TN6F, was isolated from a fruit body, and 44 strains of coexisting cultivable bacteria were isolated from its substrate, natural white rot-decayed wood. The effects of these bacteria on fungal growth were examined by an in vitro confrontation growth assay. Among the isolates, nine bacterial strains inhibited the growth of strain TN6F, while 35 strains did not affect the growth of TN6F. However, when co-cultured with strain TN6F on wood powder, many bacterial strains promoted the weight loss of the substrate. A subsequent chemical composition analysis showed that co-culturing accelerated delignification. Higher laccase activity was detected when strain TN6F was co-cultured on wood powder medium with bacterial strains TN6W-26 or TN6W-27. These results indicate that some bacterial strains might promote wood degradation.



Similar content being viewed by others
References
Baldrian P (2004) Increase of laccase activity during interspecific interactions of white-rot fungi. FEMS Microbiol Ecol 50:245–253
Blanchette RA, Shaw CG (1978) Associations among bacteria, yeasts, and basidiomycetes during wood decay. Phytopathology 68:631–637
Bontemps C, Toussaint M, Revol P-V, Hotel L, Jeanbille M, Uroz S, Turpault M-P, Blaudez D, Leblond P (2013) Taxonomic and functional diversity of Streptomyces in a forest soil. FEMS Microbiol Lett 342:157–167
Cespedes R, Salas L, Calderon I, Gonzales B, Vicuna R (1992) Microbial and biochemical-characterization of a bacterial consortium isolated from decaying wood by growth on a β-O-4 lignin-related dimeric compound. Arch Microbiol 158:162–170
Clausen CA (1996) Bacterial association with decaying wood: a review. Int Biodeter Biodegrad 37:101–107
Hailei W, Guangli Y, ** L, Yanchang G, Jun L, Guosheng L, Jianming Y (2009) Overproduction of Trametes versicolor laccase by making glucose starvation using yeast. Enzyme Microb Technol 45:146–149
Hiraishi A (1992) Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol 15:210–213
Hiscox J, Baldrian P, Rogers HJ, Boddy L (2010) Changes in oxidative enzyme activity during interspecific mycelial interactions involving the white-rot fungus Trametes versicolor. Fungal Genet Biol 47:562–571
Kamei I, Sonoki S, Haraguchi K, Kondo R (2006) Fungal bioconversion of toxic polychlorinated biphenyls by white-rot fungus, Phlebia brevispora. Appl Microbiol Biotechnol 73:932–940
Kamei I, Suhara H, Kondo R (2005) Phylogenetical approach to isolation of white-rot fungi capable of degrading polychlorinated dibenzo-p-dioxin. Appl Microbiol Biotechnol 69:358–366
Kamei I, Takagi K, Kondo R (2011) Degradation of endosulfan and endosulfan sulfate by white-rot fungus Trametes hirsute. J Wood Sci 57:317–322
Kamei I, Yoshida T, Enami D, Meguro S (2012) Coexisting Curtobacterium bacterium promotes growth of white-rot fungus Stereum sp. Curr Microbiol 64:173–178
Kirk TK, Fenn P (1982) Formation and action of the ligninolytic system in basidiomycetes. In: Frankland JC, Hedger JN, Swift MJ (eds) Decomposer basidiomycetes: their biology and ecology. Cambridge University Press, New York, pp 67–90
Line MA (1990) Identification of nitrogen-fixing enterobacteria from living Sassafras (Atherosperma moschatum Labill.) trees. Plant Soil 125:149–152
Mikluscak M, Dawson-Andoh BE (2004) Microbial colonizer of freshly sawn yellow-poplar (Liriodendron tulipifera L.) lumber in two seasons: part 2. Bacteria. Holzforschung 58:182–188
Murray AC, Woodward S (2007) Temporal change in function diversity of culturable bacteria populations in Sitka spruce stumps. For Pathol 37:217–235
Potrikus CJ, Breznak JA (1977) Nitrogen-fixing Enterobacter agglomerans isolated from guts of wood-eating termites. Appl Environ Microbiol 33:392–399
Spano SD, Jurgensen MF, Larsen MJ, Harvey AE (1982) Nitrogen-fixing bacteria in Douglas-fir residue decayed by Fomitopsis pinicola. Plant Soil 68:117–123
Valaskova V, de Boer W, Gunnewiek PJ, Pospisek M, Baldrian P (2009) Phylogenetic composition and properties of bacteria coexisting with the fungus Hypholoma fasciculare in decaying wood. ISME J 3:1221–1281
Wariishi H, Valli K, Gold MH (1992) Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium-kinetic mechanism and role of chelators. J Biol Chem 267:23688–23695
Weißhaupt P, Pritzkow W, Noll M (2012) Nitrogen sources of Oligoporus placenta and Trametes versicolor evaluated in a 23 experimental plan. Fungal Biol 116:81–89
Weißhaupt P, Pritzkow W, Noll M (2011) Nitrogen metabolism of wood decomposing basidiomycetes and their interaction with diazotrophs as revealed by IRMS. Int J Mass Spectrom 307:225–231
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Zhang HB, Yang MX, Tu R (2008) Unexpected high bacterial diversity in decaying wood of conifer as revealed by molecular method. Int Biodeter Biodegrad 62:471–474
Acknowledgements
The author would like to thank Mai Komatsuo, Ryuji Hori, and Kiyomatsu Ryohei for technical assistance with the experiments. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (Grant Nos. 23688041 and 15K14905).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kamei, I. Co-culturing Effects of Coexisting Bacteria on Wood Degradation by Trametes versicolor . Curr Microbiol 74, 125–131 (2017). https://doi.org/10.1007/s00284-016-1162-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-016-1162-1