Abstract
Anaerobic wastewater treatment offers several advantages; however, the effluent of anaerobic digesters still contains high levels of ammonium and dissolved methane that need to be removed before these effluents can be discharged to surface waters. The simultaneous anaerobic removal of methane and ammonium by denitrifying (N-damo) methanotrophs in combination with anaerobic ammonium-oxidizing (anammox) bacteria could be a potential solution to this challenge. After a molecular survey of a wastewater plant treating brewery effluent, indicating the presence of both N-damo and anammox bacteria, we started an anaerobic bioreactor with a continuous supply of methane, ammonium, and nitrite to enrich these anaerobic microorganisms. After 14 months of operation, a stable enrichment culture containing two types of ‘Candidatus Methylomirabilis oxyfera’ bacteria and two strains of ‘Ca. Brocadia’-like anammox bacteria was achieved. In this community, anammox bacteria converted 80% of the nitrite with ammonium, while ‘Ca. Methylomirabilis’ contributed to 20% of the nitrite consumption. The analysis of metagenomic 16S rRNA reads and fluorescence in situ hybridization (FISH) correlated well and showed that, after 14 months, ‘Ca. Methylomirabilis’ and anammox bacteria constituted approximately 30 and 20% of the total microbial community. In addition, a substantial part (10%) of the community consisted of Phycisphaera-related planctomycetes. Assembly and binning of the metagenomic sequences resulted in high-quality draft genome of two ‘Ca. Methylomirabilis’ species containing the marker genes pmoCAB, xoxF, and nirS and putative NO dismutase genes. The anammox draft genomes most closely related to ‘Ca. Brocadia fulgida’ included the marker genes hzsABC, hao, and hdh. Whole-reactor and batch anaerobic activity measurements with methane, ammonium, nitrite, and nitrate revealed an average anaerobic methane oxidation rate of 0.12 mmol h−1 L−1 and ammonium oxidation rate of 0.5 mmol h−1 L−1. Together, this study describes the enrichment and draft genomes of anaerobic methanotrophs from a brewery wastewater treatment plant, where these organisms together with anammox bacteria can contribute significantly to the removal of methane and ammonium in a more sustainable way.
Key points
• An enrichment culture containing both N-damo and anammox bacteria was obtained.
• Simultaneous consumption of ammonia, nitrite, and methane under anoxic conditions.
• In-depth metagenomic biodiversity analysis of inoculum and enrichment culture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Anaerobic treatment systems offer many advantages such as biogas production in the form of methane and low sludge production. However, the effluents of anaerobic treatment systems still contain dissolved methane and ammonium that need to be removed. While there are several processes that can be used to remove ammonium from the anaerobic effluents, currently there are no established biological methods to remove dissolved methane, resulting in the release of this potent greenhouse gas into the atmosphere (van Kessel et al. 2018; ** to contigs were reassembled in CLC (word size 30, bubble size 5000). The completeness of the draft genomes and contamination were assessed by CheckM (Parks et al. 2015). BLAST was used to search for key genes in the ‘Ca. Methylomirabilis’ and anammox genomes. The contigs containing marker genes were manually curated. The curated contigs were checked using the visualization and annotation tool Artemis (Rutherford et al. 2000).
Results
Performance Lieshout WWTP
The carrousel of the WWTP Lieshout is a low-loaded system that receives on average 4700 m3 effluent per day from 3 UASB reactors. The total nitrogen supply to the carrousel is about 216 kg N per day and the average ammonium and nitrate concentrations of the UASB effluent are 5.4 and 0.9 mg N L−1, respectively. After treatment in the carrousel, the ammonium and nitrate concentrations are 0.7 and 1.2 mg N L−1, indicating presence of both nitrifying and denitrifying microorganisms. Therefore, the potential rates for ammonium, nitrite, and nitrate conversions were measured in batch tests (Table 3). The 4.7 mg N L−1 ammonium removal in the carrousel results in a removal rate of about 0.33 mmol NH4+ L−1 day−1 and is in good agreement with the potential ammonium-oxidation (0.26–0.3 mmol NH4+ L−1 day−1) measured in the batch tests. The nitrite-oxidation and nitrate reduction potential exceed these values 2- to 5-fold, indicating that aerobic ammonium oxidation might be the limiting step in the nitrogen removal in WWTP Lieshout. N-damo and anammox activity could not be detected within 36 h of incubation, possibly because of high back ground denitrification rates (Table 3).
Molecular survey Lieshout sludge
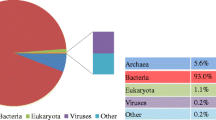
DNA of the Lieshout sludge was extracted and sequenced using Ion Torrent Technology. Extraction of 16S rRNA gene sequences (Fig. 1a) of the total metagenome yielded 2801 reads, the majority belonging to Proteobacteria (38%), Bacteroidetes (13%), Chloroflexi (12%), and Firmicutes (10%). Planctomycetes comprised 6% of the 16S rRNA reads, of which many corresponded to the order of Phycisphaerales. In the metagenome, no 16S rRNA reads belonging to anammox bacteria were retrieved, while 23 (0.8%) reads were affiliated to Nitrosomonas and 25 (0.9%) to aerobic methanotrophs (Methylococcocaea). Despite the absence of anammox reads in the metagenome, 16S rRNA and hydrazine synthase (hzsA) genes could readily be amplified using DNA from the Lieshout carrousel sludge as a template. The majority of anammox clones were affiliated to ‘Ca. Brocadia’ (Supplementary Fig. S1). Also, the pmoA gene of ‘Ca. Methylomirabilis’ could be amplified from the sludge DNA, and the diversity of these N-damo bacteria (Supplementary Fig. S2) was similar to previous studies based on nested 16S rRNA gene amplifications (Luesken et al. 2011c). The amplification of the mcrA gene showed a large diversity of methanogens (Supplementary Fig. S3) probably seeded into the carrousel from the UASB effluent. One of the 16 analyzed mcrA clones was affiliated to N-damo ‘Ca. Methanoperedens’ archaea.
Enrichment of N-damo and anammox bacteria
As anammox and N-damo bacteria were present in relatively low abundance, the excess biomass in the sludge needed to be mineralized as a strategy to reduce enrichment times. The first 3 days after inoculation of an anaerobic SBR, air was introduced into the system to convert easily degradable carbon and biomass. After the dissolved oxygen concentration started to increase, the air supply was turned off, and the SBR was fed with methane, ammonium, nitrate, and nitrite under anoxic conditions. After 100 days, the N-load to the reactor could gradually be increased to 5 mM NH4+, 8 mM NO2−, and 5 mM NO3− resulting in nitrite and ammonium consumption rates of 1.25 mmol L−1 day−1 and 1 mmol L−1 day−1, respectively (Fig. 2). When the reactor was run in batch mode without ammonium present, nitrite consumption (0.12 mmol h−1) at the expense of methane could readily be observed (Fig. 3a). In the absence of methane, anammox bacteria converted nitrite (0.5 mmol h−1) with ammonium as electron donor (Fig. 3b), indicating that anammox converted about 80% of the nitrite fed to the reactor while N-damo was responsible for the remaining 20%.
The fate of 15N-labeled substrates was measured in a reactor batch test. According to expectation (Ettwig et al. 2010), labeled 15N-nitrite was predominantly converted to 30N2 gas (Fig. 4a) by the N-damo bacteria. In the anammox test, most of the 15N label of ammonium was converted in to 29N2 following the described stoichiometry (Strous et al. 1999b). In both incubation, some 28N2 from unlabeled nitrate was produced presumably via background denitrification, and therefore exact mass balanced could not be made.
a Labeled dinitrogen gas production from 15N nitrite with methane as electron donor by N-damo, without presence of ammonium. b Labeled dinitrogen gas production from 15N ammonium with nitrite by anammox, without methane present, in the Lieshout N-damo and anammox enrichment culture after 42 months of operation
In accordance with the nitrogen conversions in reactor batch tests, ‘Ca. Methylomirabilis’ and anammox bacteria were the dominant members of the community as estimated from FISH microscopy (Fig. 5) and metagenomic analysis after 14 months of operation.
FISH microscopy of the N-damo and anammox enrichment. a After 200 days, anammox bacteria were shown to be present (amx820, pink) whereas N-damo bacteria could not be visualized by FISH (damo1027, green). b After 408 days of enrichment, both anammox bacteria (amx820, blue) and N-damo bacteria (damo1027, pink) were clearly present. Scale bar represents 20 μm
Metagenomic analysis and classification based on the 16S rRNA gene
After 4.5 months of enrichment, genomic DNA was extracted and sequenced by Ion Torrent Technology to investigate enrichment of anaerobic ammonia and methane-oxidizing microorganisms. Not much changes were observed compared with the original sludge and we decided to continue the enrichment. After 14 months of enrichment, DNA was extracted once more to analyze the 16S rRNA gene diversity and to potentially assemble draft genomes. From this metagenome, 1996 16S rRNA reads were extracted. The phylogenetic classifications for groups with an abundance of greater than 1% of the total number of 16S rRNA gene reads are shown in Fig. 1b. More than 60% of the 16S rRNA gene reads belonged to three dominant groups: Brocadiaceae (21%), N-damo ‘Ca. Methylomirabilis’ bacteria (32%), and Phycisphaera-like Planctomycetes (10%), confirming the results of FISH microscopy.
The Ion Torrent reads were assembled into contigs, and contigs were binned by differential coverage and GC content. The binning resulted in 6 high-quality draft genomes (above 92% completeness; Table 4). Two draft genomes (Bins 1 and 2) represented distinct ‘Ca. Methylomirabilis’ species, closely related to ‘Ca. M. oxyfera’ (Bin2) and ‘Ca. M. lanthanidiphila’ (Bin1). Three bins showed high similarity to ‘Ca. Brocadia’ anammox species and further refinement was hampered by microdiversity differences. Interestingly, one draft genome (Bin6) represented an unknown genus, most closely related to the Phycisphaera species within the phylum Planctomycetes.
The ‘Ca. Methylomirabilis‘ draft genomes (Bins 1 and 2) contained the diagnostic pmoCAB, xoxF, nirS, and putative NO dismutase genes. The pmoA marker genes encoding a subunit of the membrane-bound methane monooxygenase had 99.2% and 97.1% identity at the protein level to ‘Ca. M. oxyfera’ and ‘Ca. M. lanthanidiphila,’ respectively. We also identified a gene encoding the lanthanide-dependent methanol dehydrogenase (xoxF) in both genomes with 93–98% identity at the protein level to ‘Ca. Methylomirabilis’ species. Genes encoding the canonical calcium-dependent methanol dehydrogenase were absent in both genomes. Genes encoding the putative nitric oxide dismutase (nod) showed 85–90% amino acid identity to ‘Ca. Methylomirabilis’ species. The identified cd1 cytochrome nitrite reductase (nirS) had 92–99% identity to other ‘Ca. Methylomirabilis’ species.
The genomes assembled in Bins 3, 5, and 8 were determined to belong to the Planctomycetes phylum by the CheckM taxonomy assessment and most closely related to the anammox bacterium ‘Ca. Brocadia fulgida.’ This points to microdiversity of this species with bin3 as the most dominant (Supplementary Fig. S4). The genes encoding the crucial proteins involved in the anammox process were identified in the bins. These proteins include the hydrazine synthase, the hydrazine dehydrogenase, and multiple hydroxylamine oxidoreductases. The phylogenetic functional marker genes hzsA (Harhangi et al. 2012) encoding the alpha subunit of the hydrazine synthase had 89–93% identity on protein level to ‘Ca. Brocadia’ species.
The genome present in Bin6 was the only bin with complete 16S rRNA and 23S rRNA genes on one of the contigs. A BlastN search with the 16S rRNA gene revealed 99.3% identity with clones from anammox reactors treating wastewater (NCBI, unpublished sequences). The closest cultured representative with 92.4% identity was Phycisphaerae bacterium RAS2 isolated from a fish tank biofilter enrichment (Wiegand et al. 2020). The Prokka annotated genome does not reveal the potential for an autotrophic life style.
Discussion
Nitrate- and nitrite-dependent anaerobic oxidation of methane (N-damo) was discovered a decade ago, but the ecophysiological characterization of the microorganisms involved has been hindered by the slow growth of the responsible organisms (Raghoebarsing et al. 2006; Ettwig et al. 2010; Haroon et al. 2013). The N-damo microorganisms ‘Candidatus Methanoperedens nitroreducens’ archaea and ‘Ca. Methylomirabilis’ bacteria have been detected in various freshwater sediments (Welte et al. 2016). In this study, we started an enrichment culture fed with methane, ammonium, nitrate, and nitrite using carrousel sludge from a WWTP treating brewery effluent, as the inoculum. Based on activity measurements, FISH microscopy, and metagenome analyses after 14 months, the enrichment was dominated by ‘Ca. Methylomirabilis’ species and anammox bacteria. Previous enrichments were fed with mixtures of nitrite and nitrate without ammonium, to ensure that anammox bacteria would not outcompete ‘Ca. Methylomirabilis’ bacteria for nitrite (Luesken et al. 2011b; Shi et al. 2013; Hu et al. 2015; Ding et al. 2017; Vaksmaa et al. 2017b). Although the inoculum sludge had no detectable anammox or N-damo activity within 36 h, and only one 16S rRNA gene read resembling anammox could be detected in the initial metagenome, the diagnostic genes hzsA and pmoA could be readily amplified using DNA extracted from the sludge as a template. This indicated that the Lieshout carrousel sludge does harbor indigenous populations of N-damo and anammox bacteria and could serve as start-up material when large-scale installation need to be seeded (van Kessel et al. 2018). Indeed, after a start-up phase of about 100 days, an increasing consumption of nitrite, methane, and ammonium could be observed, and ultimately anammox and N-damo bacteria were the key players of the microbial community. The start-up phase was relatively short, as previous enrichments obtained from a minerotrophic peatland only showed significant methane oxidation rates after 9 months and the original enrichment of N-damo took more than a year before activity could be measured (Raghoebarsing et al. 2006; Zhu et al. 2012). In addition to substrate preference and availability, temperature has been implicated as a decisive factor in the outcome of N-damo enrichments. In enrichments started from wastewater treatment sludge and lake sediments, a co-enrichment of ‘Ca. Methylomirabilis’ bacteria and ‘Ca. Methanoperedens’ archaea was obtained at 35 °C, whereas at 22 °C, only N-damo bacteria were enriched (Hu et al. 2009). Furthermore, ‘Ca. Methanoperedens’ might be more sensitive to oxygen exposure (Guerrero-Cruz et al. 2018) than anammox and ‘Ca. Methylomirabilis’ bacteria (Luesken et al. 2011a, 2012). As we used air in the first 3 days of enrichment to degrade excess sludge, we may have strongly inhibited the few ‘Ca. Methanoperedens’ cells present and prevented their subsequent enrichment.
In the present enrichment, the cell numbers estimated by FISH correlated well with the metagenome sequencing results for ‘Ca. Methylomirabilis,’ with an abundance of about 30–40%. Anammox seemed somewhat underrepresented in the metagenome (20%) as activity measurements indicated that 80% of the nitrite conversion was accounted for by anammox activity. A similar observation was made previously where anammox converted about 70% of nitrite while FISH counts of anammox were less than 50% of the community (Luesken et al. 2011a). In our enrichment, there was always ammonium-limiting to ensure that ‘Ca. Methylomirabilis’ was not outcompeted by anammox (Hu et al. 2015). Incubations performed with the whole bioreactor revealed average ammonium oxidation and nitrite reduction after 14 months of 1 mmol day−1 L−1 and 1.25 mmol day−1 L−1, respectively which is a little bit lower compared with other enrichments (Luesken et al. 2011b; Hu et al. 2009; Vaksmaa et al. 2017b). With 15N isotope labeling experiments, we could show that in the absence of ammonium, a good methane-dependent nitrite conversion took place, and that the dominant product was the expected 30N2 gas (Ettwig et al. 2010). In the absence of methane, 15N-labeled ammonium was combined with 14N nitrite to form 29N2 by anammox bacteria in accordance with the anammox reaction equation (Strous et al. 1999b). As not all unlabeled nitrate could be removed from the reactor, some background denitrification and N2O production was observed as well, as has been reported for other N-damo enrichments (Vaksmaa et al. 2017b).
Metagenome analysis revealed that ‘Ca. Methylomirabilis,’ ‘Ca. Brocadia,’ and a Phycisphaera-like bacterium were the dominant members of the community after 14 months of enrichments. The diagnostic genes for nitrite-driven anaerobic ammonium and methane oxidation were identified in the genomes assembled from the metagenome sequences. To elucidate the role of the Phycisphaera-like bacterium, further research will be needed. Recently, members of the family Phycisphaeraceae have been reported to be potentially involved in anaerobic ammonium oxidation coupled to the reduction of sulfate (Sulfammox) or Fe(III) reduction (Feammox) in marine sediments (Rios-Del Toro et al. 2018a). Furthermore, representatives of this family were dominant in marine sediments from the eastern tropical North Pacific coast (Rios-Del Toro et al. 2018b). In this study, anaerobic ammonium oxidation linked to the microbial reduction of natural organic matter (NOM) fueled nitrogen loss. Other community members in our reactor system, mostly less than 5% of the population, belonged to various groups that have been reported before to be present in anaerobic methanotrophic enrichments (Vaksmaa et al. 2017b). In the enrichment, betaproteobacterial Rhodocyclaceae accounted for 6% and Comamonadaceae for 4% of the genome reads. They have both been implicated to perform denitrification, which may explain the observed back ground nitrate reduction, which was higher than expected based on the methane oxidation rate alone. Further significant numbers of Chloroflexi were present. They are obligate anaerobes that have previously been found in both anaerobic methanotrophic (Ettwig et al. 2009; Siniscalchi et al. 2015) and methanogenic enrichment cultures (Yamada et al. 2005; Gray et al. 2011; Liang et al. 2015). These Chloroflexi may degrade organic acids to formate, acetate, and hydrogen and feed the other community members (Hug et al. 2013).
In summary, we have enriched a N-damo and anammox coculture from the Lieshout WWTP carrousel sludge. Metagenome analysis, FISH microscopy, and activity test were in good agreement with each other that anammox contributed most to the nitrite conversion under ammonium limitation. The newly enriched co-culture will be used in future studies to unravel the ecophysiological properties of N-damo and anammox under oxygen limitation and investigate their potential role in more sustainable wastewater treatment systems.
Data availability
The raw reads of the metagenome sequencing are submitted to the Sequence Read Archive (SRA) under project number PRJEB37137.
References
Amann RI, Krumholz J, Stahl DA (1990) Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 172:762–770
Arshad A, Speth DR, de Graaf RM, Op den Camp HJM, Jetten MSM, Welte CU (2015) A metagenomics-based metabolic model of nitrate-dependent anaerobic oxidation of methane by Methanoperedens-like Archaea. Front Microbiol 6:1423
Arshad A, Dalcin Martins P, Frank J, Jetten MSM, Op den Camp HJM, Welte CU (2017) Mimicking microbial interactions under nitrate-reducing conditions in an anoxic bioreactor: enrichment of novel Nitrospirae bacteria distantly related to Thermodesulfovibrio. Environ Microbiol 19(12):4965–4977
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Bhattacharjee AS, Motlagh AM, Jetten MSM, Goel R (2016) Methane dependent denitrification- from ecosystem to laboratory-scale enrichment for engineering applications. Water Res 99:244–252
Cogert KI, Ziels RM, Winkler MKH (2019) Reducing cost and environmental impact of wastewater treatment with denitrifying methanotrophs, anammox, and mainstream anaerobic treatment. Environ Sci Technol 53(21):12935–12944
Daims H, Brühl A, Amann RI, Schleifer KH, Wagner M (1999) The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol 22:434–444
Ding ZW, Lu YZ, Fu L, Ding J, Zeng RJ (2017) Simultaneous enrichment of denitrifying anaerobic methane-oxidizing microorganisms and anammox bacteria in a hollow-fiber membrane biofilm reactor. Appl Microbiol Biotechnol 101:437–446
Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJM, Jetten MSM, Strous M (2008) Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10(11):3164–3173
Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M (2009) Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75(11):3656–3662
Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJCT, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M (2010) Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464(7288):543–548
Ettwig KF, Zhu B, Speth DR, Keltjens JT, Jetten MSM, Kartal B (2016) Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113(45):12792–12796
Fan SQ, **e GJ, Lu Y, Liu BF, **ng DF, Han HJ, Yuan Z, Ren NQ (2020) Granular sludge coupling nitrate/nitrite dependent anaerobic methane oxidation with anammox: from proof-of-concept to high rate nitrogen removal. Environ Sci Technol 54(1):297–305
Fu L, Zhang F, Bai YN, Lu YZ, Ding J, Zhou D, Liu Y, Zeng RJ (2019) Mass transfer affects reactor performance, microbial morphology, and community succession in the methane-dependent denitrification and anaerobic ammonium oxidation co-culture. Sci Total Environ 651(1):291–297
Gambelli L, Guerrero-Cruz S, Mesman RJ, Cremers G, Jetten MSM, Op den Camp HJM, Kartal B, Lueke C, van Niftrik L (2018) Community composition and ultrastructure of a nitrate-dependent anaerobic methane-oxidizing enrichment culture. Appl Environ Microbiol 84(3):e02186–e02117
Guerrero-Cruz S, Cremers G, van Alen TA, Op den Camp HJM, Jetten MSM, Rasigraf O, Vaksmaa A (2018) Response of the anaerobic methanotroph “Candidatus Methanoperedens nitroreducens” to oxygen stress. Appl Environ Microbiol 84(24):e01832–e01818
Guerrero-Cruz S, Stultiens K, van Kessel MAHJ, Versantvoort W, Jetten MSM, Op den Camp HJM, Kartal B (2019) Key physiology of a nitrite-dependent methane-oxidizing enrichment culture. Appl Environ Microbiol 85:e00124–e00119
Harhangi HR, Le Roy M, van Alen T, Hu BL, Groen J, Kartal B, Tringe SG, Quan ZX, Jetten MSM, Op den Camp HJM (2012) Hydrazine synthase, a unique phylomarker with which to study the presence and biodiversity of anammox bacteria. Appl Environ Microbiol 78(3):752–758
Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW (2013) Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500(7464):567–570
Hauck M, Maalcke-Luesken FA, Jetten MSM, Huijbregts MAJ (2016) Removing nitrogen from wastewater with side stream anammox: what are the treat-offs between environmental impacts? Resour Conserv Recycl 107:212–219
Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z (2009) Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ Microbiol Rep 1(5):377–384
Hu S, Zeng RJ, Haroon MF, Keller J, Lant PA, Tyson GW, Yuan Z (2015) A laboratory investigation of interactions between denitrifying anaerobic methane oxidation (DAMO) and anammox processes in anoxic environments. Sci Reports 5:8706
Hu Z, Wessels HJCT, van Alen TA, Jetten MSM, Kartal B (2019) Nitric oxide-dependent ammonium oxidation. Nat Commu 10:1244
Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR, Williams KH, Tringe SG, Banfield JF (2013) Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1:22
in’t Zandt MH, de Jong AE, Slomp CP, Jetten MSM (2018) The hunt for the most-wanted chemolitho-autotrophic spookmicrobes. FEMS Microbiol Ecol 94(6):fiy064
Kartal B, Kuenen JG, van Loosdrecht MCM (2010) Sewage treatment with anammox. Science 328(5979):702–703
Kartal B, Maalcke WJ, de Almeida NM, Cirpus I, Gloerich J, Geerts W, Op den Camp HJM, Harhangi HR, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Keltjens JT, Jetten MSM, Strous M (2011) Molecular mechanism of anaerobic ammonium oxidation. Nature 479(7371):127–130
Kowalchuk GA, de Bruijn FJ, Head IM, Akkermans AD, van Elsas JD (2004) Molecular microbial ecology manual (MMEM), 2nd ed, vol 1. Kluwer Academic Publishing, London, United Kingdom
Lackner S, Bitter M, Vlaeminck SE, Joss A, Horn H, van Loosdrecht MCM (2014) Full-scale partial nitritation/anammox experiences – an application survey. Water Res 55:292–303
Liu T, Hu S, Yuan Z, Guo J (2019) High-level nitrogen removal by simultaneous partial nitration, anammox and nitrite/nitrate-dependent anaerobic methane oxidation. Water Res 166:115057
Luesken FA, Sánchez J, van Alen TA, Sanabria J, Op den Camp HJM, Jetten MSM, Kartal B (2011a) Simultaneous nitrite-dependent anaerobic methane and ammonium oxidation processes. Appl Environ Microbiol 77:6802–6807
Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TL, Zeeman G, Temmink H, Strous M, Op den Camp HJM, Jetten MSM (2011b) Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol 92(4):845–854
Luesken FA, Zhu B, van Alen TA, Butler MK, Diaz MR, Song B, Op den Camp HJM, Jetten MSM, Ettwig KF (2011c) pmoA primers for detection of anaerobic methanotrophs. Appl Environ Microbiol 77(11):3877–3880
Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MSM (2012) Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ Microbiol 14(4):1024–1034
Meng H, Wang YF, Chan HW, Wu RN, Gu JD (2016) Co-occurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in subtropical acidic forest soils. Appl Microbiol Biotechnol 100(17):7727–7739
Nie WB, **e GJ, Ding J, Lu Y, Liu BF, **ng DF, Wang Q, Han HJ, Yuan Z, Ren NQ (2019) High performance nitrogen removal through integrating denitrifying anaerobic methane oxidation and anammox: from enrichment to application. Environ Int 132:105107
Nie WB, **e GJ, Ding J, Peng L, Lu Y, Tan X, Yue H, Liu BF, **ng DF, Meng J, Han HJ, Ren NQ (2020) Operation strategies of n-DAMO and anammox process based on microbial interactions for high rate nitrogen removal from landfill leachate. Environ Int 139:105596
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25(7):1043–1055
Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJP, Ettwig KF, Rijpstra WIC, Schouten S, Damste JSS, Op den Camp HJM, Jetten MSM, Strous M (2006) A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440(7086):918–921
Rios-Del Toro EE, Valenzuela,EI, López-Lozano NG, Cortés-Martínez MG, Sánchez-Rodríguez MA, Calvario-Martínez O, Sánchez-Carrillo S, Cervantes FJ (2018a) Anaerobic ammonium oxidation linked to sulfate and ferric iron reduction fuels nitrogen loss in marine sediments. Biodegradation 29(5):429–442
Rios-Del Toro EE, Valenzuela EI, Ramírez JE, López-Lozano NE, Cervantes FJ (2018b) Anaerobic ammonium oxidation linked to microbial reduction of natural organic matter in marine sediments. Environ Sci Technol Lett 5(9):571–577
RStudio Team (2015) RStudio: integrated development environment for r. RStudio, Inc., Boston, MA Retrieved from http://www.rstudio.com/
Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B (2000) Artemis: sequence visualization and annotation. Bioinformatics 16(10):944–945
Schmid MC, Schmitz-Esser S, Jetten MSM, Wagner M (2001) 16S-23S rDNA intergenic spacer and 23S rDNA of anaerobic ammonium-oxidizing bacteria: implications for phylogeny and in situ detection. Environ Microbiol 3:450–459
Shen LD, Liu S, Huang Q, Lian X, He ZF, Geng S, ** RC, He YF, Lou LP, Xu XY, Zheng P, Hu BL (2014) Evidence for the cooccurrence of nitrite-dependent anaerobic ammonium and methane oxidation processes in a flooded paddy field. Appl Environ Microbiol 80(24):7611–7619
Shi Y, Hu S, Lou J, Lu P, Keller J, Yuan Z (2013) Nitrogen removal from wastewater by coupling anammox and methane-dependent denitrification in a membrane biofilm reactor. Environ Sci Technol 47(20):11577–11583
Siniscalchi LA, Vale IC, Dell’Isola J, Chernicharo CA, Calabria Araujo J (2015) Enrichment and activity of methanotrophic microorganisms from municipal wastewater sludge. Environ Technol 36(9–12):1563–1575
Strous M, Jetten MSM (2004) Anaerobic oxidation of methane and ammonium. Annu Rev Microbiol 58:99–117
Strous M, Heijnen JJ, Kuenen JG, Jetten MSM (1998) The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol 50:589–596
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999a) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Strous M, Kuenen JG, Jetten MSM (1999b) Key physiology of anaerobic ammonium oxidation. Appl Environ Microbiol 65(7):3248–3250
Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Médigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJM, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens JT, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MSM, Wagner M, Le Paslier D (2006) Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440(7085):790–794
Stultiens K, Guerrero-Cruz S, van Kessel MAHJ, Jetten MSM, Kartal B, Op den Camp HJM (2019) Interactions between anaerobic ammonium- and methane-oxidizing microorganisms in a laboratory-scale sequencing batch reactor. Appl Microbiol Biotechnol 103:6783–6795
Vaksmaa A, Guerrero-Cruz S, van Alen TA, Cremers G, Ettwig KF, Lüke C, Jetten MSM (2017a) Enrichment of anaerobic nitrate-dependent methanotrophic ‘Candidatus Methanoperedens nitroreducens’ archaea from an Italian paddy field soil. Appl Microbiol Biotechnol 101(18):7075–7084
Vaksmaa A, Jetten MS, Ettwig KF, Lüke C (2017b) McrA primers for the detection and quantification of the anaerobic archaeal methanotroph ‘Candidatus Methanoperedens nitroreducens’. Appl Microbiol Biotechnol 101(4):1631–1641
van Kessel MAHJ, Stultiens K, Slegers MFW, Guerrero Cruz S, Jetten MSM, Kartal B, Op den Camp HJM (2018) Current perspectives on the application of N-damo and anammox in wastewater treatment. Curr Opin Biotechnol 50:222–227
Versantvoort W, Guerrero-Cruz S, Speth DR, Frank J, Gambelli L, Cremers G, van Alen T, Jetten MSM, Kartal B, Op den Camp HJM, Reimann J (2018) Comparative genomics of ‘Candidatus Methylomirabilis’ species and description of ‘Ca. Methylomirabilis lanthanidiphila’. Front Microbiol 9:1672
Welte CU, Rasigraf O, Vaksmaa A, Versantvoort W, Arshad A, Op den Camp HJM, Jetten MSM, Lueke C, Reimann J (2016) Nitrate- and nitrite-dependent anaerobic oxidation of methane. Environ Microbiol Reports 8:941–956
Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller R-W, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster A-K, Øvreås L, Rohde M, Galperin MY, Jogler C (2020) Cultivation and functional characterization of 79 Planctomycetes uncovers their unique biology. Nature Microbiol 5:126–140
Winkler MK, Ettwig KF, Vannecke TP, Stultiens K, Bogdan A, Kartal B, Volcke EI (2015) Modelling simultaneous anaerobic methane and ammonium removal in a granular sludge reactor. Water Res 73:323–331
**e GJ, Liu T, Cai C, Hu S, Yuan Z (2018) Achieving high-level nitrogen removal in mainstream by coupling anammox with denitrifying anaerobic methane oxidation in a membrane biofilm reactor. Water Res 131:196–204
Xu S, Lu W, Mustafa MF, Caicedo LM, Guo H, Fu X, Wang H (2017) Co-existence of anaerobic ammonium oxidation bacteria and denitrifying anaerobic methane oxidation bacteria in sewage sludge: community diversity and seasonal dynamics. Microb Ecol 74(4):832–840
Yamada T, Sekiguchi Y, Imachi H, Kamagata Y, Ohashi A, Harada H (2005) Diversity, localization, and physiological properties of filamentous microbes belonging to Chloroflexi subphylum I in mesophilic and thermophilic methanogenic sludge granules. Appl Environ Microbiol 71(11):7493–7503
Zhou J, Bruns MA, Tiedje JM (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322
Zhu B, van Dijk G, Fritz C, Smolders AJ, Pol A, Jetten MSM, Ettwig KF (2012) Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl Environ Microbiol 78(24):8657–8665
Zhu B, Bradford L, Huang S, Szalay A, Leix C, Weissbach M, Táncsics A, Drewes JE, Lueders T (2017) Unexpected diversity and high abundance of putative nitric oxide dismutase (Nod) genes in contaminated aquifers and wastewater treatment systems. Appl Environ Microbiol 83(4):e02750–e02716
Funding
This work was supported by the Netherlands organization for Scientific Research, SIAM 024002002 and the European Research Council [ERC AG 339880 Eco_MoM] to MSMJ, and by the European Research Council ERC AG 669371 to HJMOdC. KST, MAHJvK, and SGC were supported by STW grant 13146. MAHJvK was further supported by an NWO Veni grant (016.veni.192.062). HOdC and BK were supported by the European Research Council (Advanced Grant VOLCANO 669371 and ERC starting grant GreenT 6400422).
Author information
Authors and Affiliations
Contributions
KS, MAHJvK, BK, MSMJ, and HOdC planned research. MAHJvK, BK, and HOdC supervised the project. KS, PF, and CP executed experiments. KS, MAHJvK, and HOdC analyzed data. TAvA performed sequencing and JF helped with analyzing the sequencing data. MSMJ, HOdC, and MAHJvK wrote the manuscript. All authors discussed results and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 147 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stultiens, K., van Kessel, M.A., Frank, J. et al. Diversity, enrichment, and genomic potential of anaerobic methane- and ammonium-oxidizing microorganisms from a brewery wastewater treatment plant. Appl Microbiol Biotechnol 104, 7201–7212 (2020). https://doi.org/10.1007/s00253-020-10748-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10748-z