Abstract
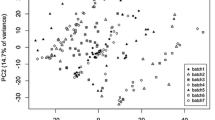
Data normalization is an essential part of a large-scale untargeted mass spectrometry metabolomics analysis. Autoscaling, Pareto scaling, range scaling, and level scaling methods for liquid chromatography-mass spectrometry data processing were compared with the most common normalization methods, including quantile normalization, probabilistic quotient normalization, and variance stabilizing normalization. These methods were tested on eight datasets from various clinical studies. The efficiency of the data normalization was assessed by the distance between clusters corresponding to batches and the distance between clusters corresponding to clinical groups in the space of principal components, as well as by the number of features with a pairwise statistically significant difference between the batches and the number of features with a pairwise statistically significant difference between clinical groups. Autoscaling demonstrated the most effective reduction in interbatch variation and can be preferable to probabilistic quotient or quantile normalization in liquid chromatography-mass spectrometry data.


Similar content being viewed by others
References
Bijlsma S, Bobeldijk I, Verheij ER, Ramaker R, Kochhar S, Macdonald IA, et al. Large-scale human metabolomics studies: a strategy for data (pre-) processing and validation. Anal Chem. 2006;78:567–74. https://doi.org/10.1021/ac051495j.
Koelmel JP, Cochran JA, Ulmer CZ, Levy AJ, Patterson RE, Olsen BC, et al. Software tool for internal standard based normalization of lipids, and effect of data-processing strategies on resulting values. BMC Bioinformatics. 2019;20:1–13. https://doi.org/10.1186/s12859-019-2803-8.
Sysi-Aho M, Katajamaa M, Yetukuri L, Orešič M. Normalization method for metabolomics data using optimal selection of multiple internal standards. BMC Bioinformatics. 2007;8:1–17. https://doi.org/10.1186/1471-2105-8-93.
De Livera AM, Sysi-Aho M, Jacob L, Gagnon-Bartsch JA, Castillo S, Simpson JA, et al. Statistical methods for handling unwanted variation in metabolomics data. Anal Chem. 2015;87:3606–15. https://doi.org/10.1021/ac502439y.
Wang SY, Kuo CH, Tseng YJ. Batch normalizer: a fast total abundance regression calibration method to simultaneously adjust batch and injection order effects in liquid chromatography/time-of-flight mass spectrometry-based metabolomics data and comparison with current calibration met. Anal Chem. 2013;85:1037–46. https://doi.org/10.1021/ac302877x.
Shen X, Gong X, Cai Y, Guo Y, Tu J, Li H, et al. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics. 2016;12:1–12. https://doi.org/10.1007/s11306-016-1026-5.
Dieterle F, Ross A, Schlotterbeck G, Senn H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in1H NMR metabonomics. Anal Chem. 2006;78:4281–90. https://doi.org/10.1021/ac051632c.
Han RH, Wang M, Fang X, Han X. Simulation of triacylglycerol ion profiles: bioinformatics for interpretation of triacylglycerol biosynthesis. J Lipid Res. 2013;54:1023–32. https://doi.org/10.1194/jlr.M033837.
van den Berg RA, Hoefsloot HCJ, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics. 2006;7:1–15. https://doi.org/10.1186/1471-2164-7-142.
Karaman I. Preprocessing and pretreatment of metabolomics data for statistical. Analysis. 2017;965:145–61. https://doi.org/10.1007/978-3-319-47656-8.
Du YM, Hu Y, **a Y, Ouyang Z. Power normalization for mass spectrometry data analysis and analytical method assessment. Anal Chem. 2016;88:3156–63. https://doi.org/10.1021/acs.analchem.5b04418.
Huber W, Von Heydebreck A, Sültmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18. https://doi.org/10.1093/bioinformatics/18.suppl_1.S96.
Lin SM, Du P, Huber W, Kibbe WA. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36:1–9. https://doi.org/10.1093/nar/gkm1075.
Chawade A, Alexandersson E, Levander F. Normalyzer: a tool for rapid evaluation of normalization methods for omics data sets. J Proteome Res. 2014;13:3114–20. https://doi.org/10.1021/pr401264n.
Li B, Tang J, Yang Q, Cui X, Li S, Chen S, et al. Performance evaluation and online realization of data-driven normalization methods used in LC/MS based untargeted metabolomics analysis. Sci Rep. 2016;6:1–13. https://doi.org/10.1038/srep38881.
Lee J, Park J, Lim MS, Seong SJ, Seo JJ, Park SM, et al. Quantile normalization approach for liquid chromatography- mass spectrometry-based metabolomic data from healthy human volunteers. Anal Sci. 2012;28:801–5. https://doi.org/10.2116/analsci.28.801.
Yang Q, Wang Y, Zhang Y, Li F, **a W, Zhou Y, et al. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020;48:W436–48. https://doi.org/10.1093/nar/gkaa258.
Chen J, Zhang P, Lv M, Guo H, Huang Y, Zhang Z, et al. Influences of normalization method on biomarker discovery in gas chromatography-mass spectrometry-based untargeted metabolomics: what should be considered? Anal Chem. 2017;89:5342–8. https://doi.org/10.1021/acs.analchem.6b05152.
Ejigu BA, Valkenborg D, Baggerman G, Vanaerschot M, Witters E, Dujardin JC, et al. Evaluation of normalization methods to pave the way towards large-scale LC-MS-based metabolomics profiling experiments. Omi A J Integr Biol. 2013;17:473–85. https://doi.org/10.1089/omi.2013.0010.
Ly-Verdú S, Gröger TM, Arteaga-Salas JM, Brandmaier S, Kahle M, Neschen S, et al. Combining metabolomic non-targeted GC×GC-ToF-MS analysis and chemometric ASCA-based study of variances to assess dietary influence on type 2 diabetes development in a mouse model. Anal Bioanal Chem. 2015;407:343–54. https://doi.org/10.1007/s00216-014-8227-4.
Li B, Tang J, Yang Q, Li S, Cui X, Li Y, et al. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017;45:W162–70. https://doi.org/10.1093/nar/gkx449.
Chagovets V, Wang Z, Kononikhin A, Starodubtseva N, Borisova A, Salimova D, et al. A comparison of tissue spray and lipid extract direct injection electrospray ionization mass spectrometry for the differentiation of Eutopic and ectopic endometrial tissues. J Am Soc Mass Spectrom. 2018;29:323–30. https://doi.org/10.1007/s13361-017-1792-y.
Tokareva AO, Chagovets VV, Rodionov VV, Kometova VV, Rodionova MV, Starodubtseva NL, Frankevich VE. Lipid markers of metastatic lesions in regional lymph nodes in patients with breast cancer. Akusherstvo i Ginekol (Russian Fed) 2020:133–40.
Starodubtseva N, Chagovets V, Borisova A, Salimova D, Aleksandrova N, Chingin K, et al. Identification of potential endometriosis biomarkers in peritoneal fluid and blood plasma via shotgun lipidomics. Clin Mass Spectrom. 2019;13:21–6. https://doi.org/10.1016/j.clinms.2019.05.007.
Kan NE, Khachatryan ZV, Chagovets VV, Starodubtseva NL, Amiraslanov EY, Tyutyunnik VL, et al. Analysis of metabolic pathways in intrauterine growth restriction. Biomeditsinskaya Khimiya. 2020;66:174–80. https://doi.org/10.18097/PBMC20206602174.
Tonoyan NM, Tokareva AO, Chagovets V V., Starodubtseva NL, Kozachenko IF, Adamyan LV. Possibilities for predicting recurrent uterine myoma by plasma lipidomic analysis. Akusherstvo i Ginekol (Russian Fed 2019:136–151.
Cook T, Ma Y, Gamagedara S. Evaluation of statistical techniques to normalize mass spectrometry-based urinary metabolomics data. J Pharm Biomed Anal. 2020;177:112854. https://doi.org/10.1016/j.jpba.2019.112854.
Acknowledgments
This work was financially supported by the Russian Science Foundation (No. 18-75-10097). The authors are grateful to the laboratory for the collection and storage of biological material (biobank) for providing tissue samples.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tokareva, A.O., Chagovets, V.V., Kononikhin, A.S. et al. Normalization methods for reducing interbatch effect without quality control samples in liquid chromatography-mass spectrometry-based studies. Anal Bioanal Chem 413, 3479–3486 (2021). https://doi.org/10.1007/s00216-021-03294-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03294-8