Abstract
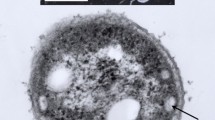
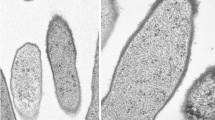
A new taxon is created for the thermophilic purple nonsulfur bacterium previously designated as Rhodopseudomonas strain GI. Strain GI was isolated from a New Mexico (USA) hot spring microbial mat and grows optimally above 40 °C and to a maximum of 47 °C. Strain GI is a bacteriochlorophyll b-containing species of purple nonsulfur bacteria and displays a budding morphology, typical of species of the genus Blastochloris. Although resembling the species Blc. viridis in many respects, the absorption spectrum, carotenoid content, and lipid fatty acid profile of strain GI is distinct from that of Blc. viridis strain DSM133T and other recognized Blastochloris species. Strain GI forms its own subclade within the Blastochloris clade of purple nonsulfur bacteria based on comparative 16S rRNA gene sequences, and its genome is significantly larger than that of strain DSM133T; average nucleotide identity between the genomes of Blc. viridis and strain GI was below 85%. Moreover, concatenated sequence analyses of PufLM and DnaK clearly showed strain GI to be distinct from both Blc. viridis and Blc. sulfoviridis. Because of its unique assortment of properties, it is proposed to classify strain GI as a new species of the genus Blastochloris, as Blc. tepida, sp.n., with strain GIT designated as the type strain (= ATCC TSD-138 = DSM 106918).





Similar content being viewed by others
References
Alberti M, Burke DH, Hearst JE (1995) Structure and sequence of the photosynthesis gene cluster. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, pp 1083–1106
Castenholz RW, Pierson BK (1995) Ecology of thermophilic anoxygenic phototrophs. In: Blankenship RE, Madigan MT, Bauer CE (eds) Anoxygenic photosynthetic bacteria. Kluwer, Dordrecht, pp 87–103
Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J (2013) Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569
Cole JK, Hutchison JR, Renslow RS, Kim Y-M, Chrisler WB, Engelmann HE, Dohnalkova AC, Hu D, Metz TO, Frederickson JK, Lindemann SR (2014) Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph–heterotroph interactions. Front Microbiol 5:109. https://doi.org/10.3389/fmicb.2014.00109
Deisenhofer J, Epp O, Miki K, Huber R, Michel H (1985) Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature 318:618–624
Drews G, Giesbrecht P (1966) Rhodopseudomonas viridis, nov. spec., ein neu isoliertes, obligat phototrophes bakterium. Arch Mikrobiol 53:255–262
Favinger J, Stadtwald R, Gest H (1989) Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Ant van Leeuwenhoek 55:291–296
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Heda GD, Madigan MT (1986) Utilization of amino acids and lack of diazotrophy in the thermophilic anoxygenic phototroph Chloroflexus aurantiacus. J Gen Microbiol 132:2469–2473
Hiraishi A (1997) Transfer of the bacteriochlorophyll b-containing phototrophic bacteria Rhodopseudomonas viridis and Rhodopseudomonas sulfoviridis to the genus Blastochloris gen. nov. Intl J Syst Bacteriol 47:217–219
Hiraishi A (2017a) Characterization of thermotolerant phototrophic bacteria, Rhodoplanes tepidicaeni sp. nov., and Rhodoplanes azumiensis sp. nov., isolated from a geothermal spring. Intl J Syst Evol Microbiol 67:5038–5045
Hiraishi A (2017b) Proposal of Rhodoplanes tepidamans sp. nov. to accommodate the thermotolerant phototrophic bacterium previously referred to as ‘Rhodoplanes (Rhodopseudomonas) cryptolactis’. Intl J Syst Evol Microbiol 67:1540–1545
Hisada T, Okamura K, Hiraishi A (2007) Isolation and characterization of phototrophic purple nonsulfur bacteria from Chloroflexus and cyanobacterial mats in hot springs. Microbes Environ 22:405–411
Hoogewerf GE, Jung DO, Madigan MT (2003) Evidence for limited species diversity of bacteriochlorophyll b-containing purple nonsulfur anoxygenic phototrophs in freshwater habitats. FEMS Microbiol Letts 218:359–364
Imhoff JF, Trüper HG, Pfennig N (1984) Rearrangement of the species and genera of phototrophic “purple nonsulfur bacteria”. Intl J Syst Bacteriol 34:340–343
Katoh K, Castresana J (2014) MAFFT: iterative refinement and additional methods. Meth Mol Biol 1079:131–146
Kempher ML, Madigan MT (2012) Phylogeny and photoheterotrophy in the acidophilic phototrophic purple bacterium Rhodoblastus acidophilus. Arch Microbiol 194:567–574
Keppen OI, Gorlenko VM (1975) A new species of purple budding bacteria containing bacteriochlorophyll b. Mikrobiologiya 44:258–264
Kompantseva EI, Imhoff JF, Thiemann B, Panteleeva EE, Akimov VN (2007) Comparative study of the fatty acid composition of some groups of purple nonsulfur bacteria. Microbiology (English translation of Mikrobiologiya) 76:541–551
Liu L-N, Faulkner M, Liu X, Huang F, Darby AC, Hall N (2016) Revised genome sequence of the purple photosynthetic bacterium Blastochloris viridis. Genome Announcements 4:e10152. https://doi.org/10.1128/genomeA.01520-15
Madigan MT (1984) A novel photosynthetic purple bacterium isolated from a Yellowstone hot spring. Science 225:313–315
Madigan MT (1986) Chromatium tepidum, sp.n., a thermophilic photosynthetic bacterium of the family Chromatiaceae. Intl J Syst Bacteriol 36:222–227
Madigan MT, Jung DO (2009) An overview of purple bacteria: systematics, physiology, and habitats. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT (eds) The purple phototrophic bacteria. Springer, Dordrecht, pp 1–15
Michel H (1982) Three-dimensional crystals of a membrane protein complex: the photosynthetic reaction centre from Rhodopseudomonas viridis. J Mol Biol 158:567–572
Nagashima S, Nagashima KVP (2013) Comparison of photosynthesis gene clusters retrieved from total genome sequences of purple bacteria. Adv Bot Res 66:151–178
Nagatsuma S, Gotou K, Yamashita T, Yu L-J, Shen J-R, Madigan MT, Kimura Y, Wang-Otomo Z-Y (2019) Phospholipid distributions in purple phototrophic bacteria and LH1-RC core complexes. Biochim Biophys Acta 1860:461–468
Namsaraev ZB, Gorlenko VM, Namsaraev BB, Buryukhaev SP, Yurkov VV (2003) The structure and biogeochemical activity of the phototrophic communities from the Bol’sherechenskii alkaline hot spring. Microbiology (English translation of Mikrobiologiya) 72:228–238
Okamura K, Hisada T, Hiraishi A (2007) Characterization of thermotolerant purple nonsulfur bacteria isolated from hot-spring Chloroflexus mats and the reclassification of “Rhodopseudomonas cryptolactis” Stadtwald-Demchick et al. 1990 as Rhodoplanes cryptolactis nom. rev., comb. nov. J Gen Appl Microbiol 53:357–361
Ramana VV, Kapoor S, Shobha E, Ramprasad EVV, Sasikala Ch, Ramana ChV (2011) Blastochloris gulmargensis sp. nov., isolated from an epilithic phototrophic biofilm. Intl J Syst Evol Microbiol 61:1811–1816
Resnick SM, Madigan MT (1989) Isolation and characterization of a mildly thermophilic nonsulfur purple bacterium containing bacteriochlorophyll b. FEMS Microbiol Lett 65:165–170
Scheer H, Svec WA, Cope BT, Studier MH, Scott RG, Katz JJ (1974) Structure of bacteriochlorophyll b. J Am Chem Soc 96:3714–3716
Stadtwald-Demchick R, Turner FR, Gest H (1990) Rhodopseudomonas cryptolactis, sp. nov., a new thermotolerant species of budding phototrophic bacteria. FEMS Microbiol Lett 71:117–122
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313
Stepkowski T, Czaplinska M, Miedzinska K, Moulin L (2003) The variable part of the dnaK gene as an alternative marker for phylogenetic studies of rhizobia and related Alpha Proteobacteria. Syst Appl Microbiol 26:483–494
Swingley WD, Blankenship RE, Raymond J (2009) Evolutionary relationships among purple photosynthetic bacteria and the origin of proteobacterial photosynthetic systems. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT (eds) The purple phototrophic bacteria. Springer, Dordrecht, pp 17–29
Takami H, Taniguchi T, Arai W, Takemoto K, Moriya Y, Goto S (2016) An automated system for evaluation of the potential functionome: MAPLE version 2.1.0. DNA Res 23:467–475
Talavera G, Castresana J (2007) Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol 56:564–577
Tanizawa Y, Fujisawa T, Nakamura Y (2017) DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 34:1037–1039
Tank M, Thiel V, Imhoff JF (2009) Phylogenetic relationship of phototrophic purple sulfur bacteria according to pufL and pufM genes. Intl Microbiol 12:175–185
Trüper HG, Imhoff JF (1989) Genus Rhodopseudomonas. In: Staley JT, Bryant MP, Pfennig N, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 3. Williams and Wilkins, Baltimore, pp 1672–1677
Tsukatani Y, Hirose Y, Harada J, Misawa N, Mori K, Inoue K, Tamiaki H (2015) Complete genome sequence of the bacteriochlorophyll b-producing photosynthetic bacterium Blastochloris viridis. Genome Announcements 3:e1006–e1015. https://doi.org/10.1128/genomeA.01006-15
Tsukatani Y, Hirose Y, Harada J, Yonekawa C, Tamiaki H (2019) Unusual features in the photosynthetic machinery of Halorhodospira halochloris DSM 1059 revealed by complete genome sequencing. Photosynth Res 140:311–319
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963
Zengler K, Heider J, Rosselló-Mora R, Widdel F (1999) Phototrophic utilization of toluene under anoxic conditions by a new strain of Blastochloris sulfoviridis. Arch Microbiol 172:204–212
Acknowledgements
We thank Professor Aharon Oren, Hebrew University of Jerusalem, for nomenclatural advice and Dr. Ch. V. Ramana, University of Hyderabad, for cells of Blc. gulmargensis for carotenoid analyses. MTM thanks Dr. John Bauld for the mat sample from which strain GI was isolated. This work was supported in part by a grant to MTM from the NASA Exobiology Program and from JSPS KAKENHI grant number 17H05231 to YT. Electron microscopy was performed at the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by the United States Department of Energy Office of Biological and Environmental Research, located at the Pacific Northwest National Laboratory (Richland, WA).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no financial or other conflicts of interest.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Genomic accession numbers Blastochloris tepida strain GIT 16S rRNA gene sequence, MG725814; Blastochloris tepida strain GIT complete genome sequence, AP018907. Culture accession numbers: Blastochloris tepida strain GIT DSM 106918; ATCC TSD-138.
Rights and permissions
About this article
Cite this article
Madigan, M.T., Resnick, S.M., Kempher, M.L. et al. Blastochloris tepida, sp. nov., a thermophilic species of the bacteriochlorophyll b-containing genus Blastochloris. Arch Microbiol 201, 1351–1359 (2019). https://doi.org/10.1007/s00203-019-01701-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-019-01701-4