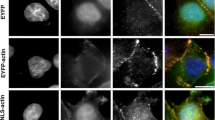
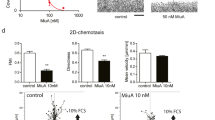
Abstract
Actin has emerged as a versatile regulator of gene transcription. Cytoplasmatic actin regulates mechanosensitive-signaling pathways such as MRTF–SRF and Hippo-YAP/TAZ. In the nucleus, both polymerized and monomeric actin directly interfere with transcription-associated molecular machineries. Natural actin-binding compounds are frequently used tools to study actin-related processes in cell biology. However, their influence on transcriptional regulation and intranuclear actin polymerization is poorly understood to date. Here, we analyze the effects of two representative actin-binding compounds, Miuraenamide A (polymerizing properties) and Latrunculin B (depolymerizing properties), on transcriptional regulation in primary cells. We find that actin stabilizing and destabilizing compounds inversely shift nuclear actin levels without a direct influence on polymerization state and intranuclear aspects of transcriptional regulation. Furthermore, we identify Miuraenamide A as a potent inducer of G-actin-dependent SRF target gene expression. In contrast, the F-actin-regulated Hippo-YAP/TAZ axis remains largely unaffected by compound-induced actin aggregation. This is due to the inability of AMOTp130 to bind to the amorphous actin aggregates resulting from treatment with miuraenamide. We conclude that actin-binding compounds predominantly regulate transcription via their influence on cytoplasmatic G-actin levels, while transcriptional processes relying on intranuclear actin polymerization or functional F-actin networks are not targeted by these compounds at tolerable doses.






Similar content being viewed by others
References
Pollard TD, Cooper JA (2009) Actin, a central player in cell shape and movement. Science 326(5957):1208–1212. https://doi.org/10.1126/science.1175862
Virtanen JA, Vartiainen MK (2017) Diverse functions for different forms of nuclear actin. Curr Opin Cell Biol 46:33–38. https://doi.org/10.1016/j.ceb.2016.12.004
Falahzadeh K, Banaei-Esfahani A, Shahhoseini M (2015) The potential roles of actin in the nucleus. Cell J 17(1):7–14
de Lanerolle P, Serebryannyy L (2011) Nuclear actin and myosins: life without filaments. Nat Cell Biol 13(11):1282–1288. https://doi.org/10.1038/ncb2364
Vartiainen MK (2008) Nuclear actin dynamics—from form to function. FEBS Lett 582(14):2033–2040. https://doi.org/10.1016/j.febslet.2008.04.010
Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR (2008) The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol 15(5):469–476. https://doi.org/10.1038/nsmb.1403
Kapoor P, Shen X (2014) Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol 24(4):238–246. https://doi.org/10.1016/j.tcb.2013.10.007
Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozak P, Grummt I (2004) Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 6(12):1165–1172. https://doi.org/10.1038/ncb1190
Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, Hozak P, de Lanerolle P (2004) Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol 6(11):1094–1101. https://doi.org/10.1038/ncb1182
Hu P, Wu S, Hernandez N (2004) A role for beta-actin in RNA polymerase III transcription. Genes Dev 18(24):3010–3015. https://doi.org/10.1101/gad.1250804
Almuzzaini B, Sarshad AA, Rahmanto AS, Hansson ML, Von Euler A, Sangfelt O, Visa N, Farrants AK, Percipalle P (2016) In beta-actin knockouts, epigenetic reprogramming and rDNA transcription inactivation lead to growth and proliferation defects. FASEB J Off Publ Fed Am Soc Exp Biolgy 30(8):2860–2873. https://doi.org/10.1096/fj.201600280R
Serebryannyy LA, Cruz CM, de Lanerolle P (2016) A role for nuclear actin in HDAC 1 and 2 regulation. Sci Rep 6:28460. https://doi.org/10.1038/srep28460
Toh KC, Ramdas NM, Shivashankar GV (2015) Actin cytoskeleton differentially alters the dynamics of lamin A, HP1alpha and H2B core histone proteins to remodel chromatin condensation state in living cells. Integr Biol (Camb). https://doi.org/10.1039/c5ib00027k
Mammoto A, Mammoto T, Ingber DE (2012) Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci 125(Pt 13):3061–3073. https://doi.org/10.1242/jcs.093005
Baarlink C, Wang H, Grosse R (2013) Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 340(6134):864–867. https://doi.org/10.1126/science.1235038
Posern G, Treisman R (2006) Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16(11):588–596. https://doi.org/10.1016/j.tcb.2006.09.008
Vartiainen MK, Guettler S, Larijani B, Treisman R (2007) Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 316(5832):1749–1752. https://doi.org/10.1126/science.1141084
Mana-Capelli S, Paramasivam M, Dutta S, McCollum D (2014) Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell 25(10):1676–1685. https://doi.org/10.1091/mbc.E13-11-0701
Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25(1):51–63. https://doi.org/10.1101/gad.2000111
Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W (2011) Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem 286(9):7018–7026. https://doi.org/10.1074/jbc.C110.212621
Gaspar P, Tapon N (2014) Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 31:74–83. https://doi.org/10.1016/j.ceb.2014.09.003
Wada K, Itoga K, Okano T, Yonemura S, Sasaki H (2011) Hippo pathway regulation by cell morphology and stress fibers. Development 138(18):3907–3914. https://doi.org/10.1242/dev.070987
Allingham JS, Klenchin VA, Rayment I (2006) Actin-targeting natural products: structures, properties and mechanisms of action. Cell Mol Life Sci CMLS 63(18):2119–2134. https://doi.org/10.1007/s00018-006-6157-9
Olson EN, Nordheim A (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11(5):353–365. https://doi.org/10.1038/nrm2890
Collin O, Na S, Chowdhury F, Hong M, Shin ME, Wang F, Wang N (2008) Self-organized podosomes are dynamic mechanosensors. Curr Biol 18(17):1288–1294. https://doi.org/10.1016/j.cub.2008.07.046
Chang CY, Leu JD, Lee YJ (2015) The actin depolymerizing factor (ADF)/cofilin signaling pathway and DNA damage responses in cancer. Int J Mol Sci 16(2):4095–4120. https://doi.org/10.3390/ijms16024095
Ojima D, Yasui A, Tohyama K, Tokuzumi K, Toriihara E, Ito K, Iwasaki A, Tomura T, Ojika M, Suenaga K (2016) Total synthesis of miuraenamides A and D. J Org Chem 81(20):9886–9894. https://doi.org/10.1021/acs.joc.6b02061
Karmann L, Schultz K, Herrmann J, Muller R, Kazmaier U (2015) Total syntheses and biological evaluation of miuraenamides. Angew Chem Int Ed Engl 54(15):4502–4507. https://doi.org/10.1002/anie.201411212
Iizuka T, Fudou R, Jojima Y, Ogawa S, Yamanaka S, Inukai Y, Ojika M (2006) Miuraenamides A and B, novel antimicrobial cyclic depsipeptides from a new slightly halophilic myxobacterium: taxonomy, production, and biological properties. J Antibiot (Tokyo) 59(7):385–391. https://doi.org/10.1038/ja.2006.55
Cheng Z, Garvin D, Paguio A, Stecha P, Wood K, Fan F (2010) Luciferase reporter assay system for deciphering GPCR pathways. Curr Chem Genom 4:84–91. https://doi.org/10.2174/1875397301004010084
Hendrix J, Baumgartel V, Schrimpf W, Ivanchenko S, Digman MA, Gratton E, Krausslich HG, Muller B, Lamb DC (2015) Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J Cell Biol 210(4):629–646. https://doi.org/10.1083/jcb.201504006
Small J, Rottner K, Hahne P, Anderson KI (1999) Visualising the actin cytoskeleton. Microsc Res Tech 47(1):3–17. https://doi.org/10.1002/(SICI)1097-0029(19991001)47:1%3c3:AID-JEMT2%3e3.0.CO;2-2
Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, Mikkelsen TS (2014) Characterization of directed differentiation by high-throughput single-cell RNA-Seq. BioRxiv. https://doi.org/10.1101/003236
Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I (2017) zUMIs: A fast and flexible pipeline to process RNA sequencing data with UMIs. BioRxiv. https://doi.org/10.1101/153940
McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ (2006) Nucleoplasmic beta-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 172(4):541–552. https://doi.org/10.1083/jcb.200507101
Wachsmuth M, Waldeck W, Langowski J (2000) Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J Mol Biol 298(4):677–689. https://doi.org/10.1006/jmbi.2000.3692
Brown CM, Dalal RB, Hebert B, Digman MA, Horwitz AR, Gratton E (2008) Raster image correlation spectroscopy (RICS) for measuring fast protein dynamics and concentrations with a commercial laser scanning confocal microscope. J Microsc 229(Pt 1):78–91. https://doi.org/10.1111/j.1365-2818.2007.01871.x
Hendrix J, Lamb DC (2014) Implementation and application of pulsed interleaved excitation for dual-color FCS and RICS. Methods Mol Biol 1076:653–682. https://doi.org/10.1007/978-1-62703-649-8_30
Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E (2005) Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J 89(2):1317–1327. https://doi.org/10.1529/biophysj.105.062836
Hendrix J, Schrimpf W, Holler M, Lamb DC (2013) Pulsed interleaved excitation fluctuation imaging. Biophys J 105(4):848–861. https://doi.org/10.1016/j.bpj.2013.05.059
Hendrix J, Dekens T, Schrimpf W, Lamb DC (2016) Arbitrary-region raster image correlation spectroscopy. Biophys J 111(8):1785–1796. https://doi.org/10.1016/j.bpj.2016.09.012
Miralles F, Posern G, Zaromytidou AI, Treisman R (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113(3):329–342
Reddy P, Deguchi M, Cheng Y, Hsueh AJ (2013) Actin cytoskeleton regulates Hippo signaling. PLoS One 8(9):e73763. https://doi.org/10.1371/journal.pone.0073763
Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R (2014) Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev 28(9):943–958. https://doi.org/10.1101/gad.239327.114
Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S (2015) Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17(9):1218–1227. https://doi.org/10.1038/ncb3216
Belin BJ, Cimini BA, Blackburn EH, Mullins RD (2013) Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell 24(7):982–994. https://doi.org/10.1091/mbc.E12-09-0685
Melak M, Plessner M, Grosse R (2017) Actin visualization at a glance. J Cell Sci. https://doi.org/10.1242/jcs.189068
Miyamoto K, Gurdon JB (2013) Transcriptional regulation and nuclear reprogramming: roles of nuclear actin and actin-binding proteins. Cell Mol Life Sci CMLS 70(18):3289–3302. https://doi.org/10.1007/s00018-012-1235-7
Rajakyla EK, Vartiainen MK (2014) Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 5:e27539. https://doi.org/10.4161/sgtp.27539
Spencer VA, Costes S, Inman JL, Xu R, Chen J, Hendzel MJ, Bissell MJ (2011) Depletion of nuclear actin is a key mediator of quiescence in epithelial cells. J Cell Sci 124(Pt 1):123–132. https://doi.org/10.1242/jcs.073197
Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK (2012) Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci USA 109(9):E544–E552. https://doi.org/10.1073/pnas.1118880109
Sharili AS, Kenny FN, Vartiainen MK, Connelly JT (2016) Nuclear actin modulates cell motility via transcriptional regulation of adhesive and cytoskeletal genes. Sci Rep 6:33893. https://doi.org/10.1038/srep33893
Stuven T, Hartmann E, Gorlich D (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 22(21):5928–5940. https://doi.org/10.1093/emboj/cdg565
Pollard TD (2016) Actin and actin-binding proteins. Cold Spring Harb Perspect Biol. https://doi.org/10.1101/cshperspect.a018226
Baarlink C, Plessner M, Sherrard A, Morita K, Misu S, Virant D, Kleinschnitz EM, Harniman R, Alibhai D, Baumeister S, Miyamoto K, Endesfelder U, Kaidi A, Grosse R (2017) A transient pool of nuclear F-actin at mitotic exit controls chromatin organization. Nat Cell Biol. https://doi.org/10.1038/ncb3641
Plessner M, Grosse R (2015) Extracellular signaling cues for nuclear actin polymerization. Eur J Cell Biol. https://doi.org/10.1016/j.ejcb.2015.05.009
Belin BJ, Lee T, Mullins RD (2015) DNA damage induces nuclear actin filament assembly by formin-2 and spire-(1/2) that promotes efficient DNA repair. Elife. https://doi.org/10.7554/eLife.07735
Matsui Y, Lai ZC (2013) Mutual regulation between Hippo signaling and actin cytoskeleton. Protein Cell 4(12):904–910. https://doi.org/10.1007/s13238-013-3084-z
Yu FX, Guan KL (2013) The Hippo pathway: regulators and regulations. Genes Dev 27(4):355–371. https://doi.org/10.1101/gad.210773.112
Dai X, She P, Chi F, Feng Y, Liu H, ** D, Zhao Y, Guo X, Jiang D, Guan KL, Zhong TP, Zhao B (2013) Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem 288(47):34041–34051. https://doi.org/10.1074/jbc.M113.518019
Isermann P, Lammerding J (2013) Nuclear mechanics and mechanotransduction in health and disease. Curr Biol 23(24):R1113–R1121. https://doi.org/10.1016/j.cub.2013.11.009
Dahl KN, Ribeiro AJ, Lammerding J (2008) Nuclear shape, mechanics, and mechanotransduction. Circ Res 102(11):1307–1318. https://doi.org/10.1161/CIRCRESAHA.108.173989
Acknowledgements
The authors thank the labs of Prof. Robert Grosse and Prof. Bin Zhao for the sharing of plasmids and fruitful scientific discussions. We, furthermore, thank Jana Peliskova for excellent technical assistance and Dr. Lisa Karmann for providing synthetic samples of Miuraenamide A. This work was funded by the Deutsche Forschungsgemeinschaft (DFG), SFB 1032, projects B08 and B03, and FOR1406.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gegenfurtner, F.A., Zisis, T., Al Danaf, N. et al. Transcriptional effects of actin-binding compounds: the cytoplasm sets the tone. Cell. Mol. Life Sci. 75, 4539–4555 (2018). https://doi.org/10.1007/s00018-018-2919-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-018-2919-4