Abstract
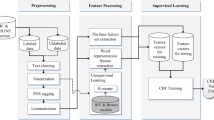
This study employs the widely used Large Language Model (LLM), BERT, to implement Named Entity Recognition (NER) on the CORD-19 biomedical literature corpus. By fine-tuning the pre-trained BERT on the CORD-NER dataset, the model gains the ability to comprehend the context and semantics of biomedical named entities. The refined model is then utilized on the CORD-19 to extract more contextually relevant and updated named entities. However, fine-tuning large datasets with LLMs poses a challenge. To counter this, two distinct sampling methodologies are proposed to apply on each dataset. First, for the NER task on the CORD-19, a Latent Dirichlet Allocation (LDA) topic modeling technique is employed. This maintains the sentence structure while concentrating on related content. Second, a straightforward greedy method is deployed to gather the most informative data of 25 entity types from the CORD-NER dataset. The study realizes its goals by demonstrating the content comprehension capability of BERT-based models without the necessity of supercomputers, and converting the document-level corpus into a source for NER data, enhancing data accessibility. The outcomes of this research can shed light on the potential progression of more sophisticated NLP applications across various sectors, including knowledge graph creation, ontology learning, and conversational AI.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Scherbakov, V., Mayorov, V.: Finetuning BERT on partially annotated NER corpora. ar**v. (2022). https://doi.org/10.48550/ar**v.2211.14360
Park, Y.I., Lee, M., Yang, G., Park, S.J., Sohn, C.: Biomedical text NER tagging tool with web interface for generating BERT-based fine-tuning dataset. Appl. Sci. 12, 12012 (2022)
Balkus, S.V., Yan, D.: Improving short text classification with augmented data using GPT-3. Ar**v, abs/2205.10981 (2022)
Kieuvongngam, V., Tan, B., Niu, Y.: Automatic text summarization of COVID-19 medical research articles using BERT and GPT-2. Ar**v, abs/2006.01997 (2020)
Maltoudoglou, L., Paisios, A., Papadopoulos, H.: BERT-based conformal predictor for sentiment analysis. In Conformal and Probabilistic Prediction and Applications, pp. 269–284. PMLR (2020)
Wang, X., Song, X., Guan, Y., Li, B., Han, J.: Comprehensive named entity recognition on CORD-19 with distant or weak supervision. Ar**v, abs/2003.12218 (2020)
Pestryakova, S., et al.: CovidPubGraph: a FAIR knowledge graph of COVID-19 publications. Sci. Data 9, 389 (2022)
Blei, D.M., Ng, A., Jordan, M.I.: Latent dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022 (2001). https://doi.org/10.1016/B978-0-12-411519-4.00006-9
Devlin, J., Chang, M., Lee, K., Toutanova, K.: BERT: pre-training of deep bidirectional transformers for language understanding. Ar**v (2019). https://doi.org/10.48550/ar**v.1810.04805
Giles, O., Huntley, R.P., Karlsson, A., Lomax, J., Malone, J.: Reference ontology and database annotation of the COVID-19 Open Research Dataset (CORD-19). bioRxiv (2020). https://doi.org/10.1101/2020.10.04.325266
Wu, J., Wang, P., Wei, X., Rajtmajer, S.M., Giles, C.L., Griffin, C.: Acknowledgement entity recognition in CORD-19 papers. In: SDP, pp. 10-19 (2020). https://doi.org/10.18653/v1/2020.sdp-1.3
Popovski, G., Kochev, S., Korousic-Seljak, B., Eftimov, T.: FoodIE: a rule-based named-entity recognition method for food information extraction. Int. Conf. Pattern Recogn. Appl. Meth. 12, 915 (2019)
Dekhili, G., Sadat, F.: Hybrid statistical and attentive deep neural approach for named entity recognition in historical newspapers. In: Conference and Labs of the Evaluation Forum (2020)
Wang, L.L., et al.: CORD-19: the COVID-19 open research dataset. Ar**v (2020)
5 Probabilistic Training Data Sampling Methods in Machine Learning. https://towardsdatascience.com/5-probabilistic-training-data-sampling-methods-in-machine-learning-460f2d6ffd9. Accessed 1 July 2023
Liu, J., et al.: Tracing the pace of COVID-19 research: topic modeling and evolution. Big Data Res. 25, 100236–100236 (2021). https://doi.org/10.1016/j.bdr.2021.100236
Unified Medical Language System(UMLS). https://www.nlm.nih.gov/research/umls/ knowledge_sources/metathesaurus/index.html. Accessed 8 July 2023
SpaCy models for biomedical text processing. https://allenai.github.io/scispacy/. Accessed 8 July 2023
David Chuan-En Lin, 8 Simple Techniques to Prevent Overfitting. https://towardsdatascience.com/8-simple-techniques-to-prevent-overfitting-4d443da2ef7d. Accessed 1 July 2023
Thant, S., Anutariya, C., Andres, F., Racharak, T.: BERT fine-tuned CORD-19 NER dataset, IEEE Dataport (2023). https://doi.org/10.21227/m7gj-ks21
ShinThant3010, ‘ShinThant3010/Deep-Learning-based-KG-for-Covid19-Vaccination: Deep Learning based KG for Covid19 Vaccination’. Zenodo, 02 November 2023. https://doi.org/10.5281/zenodo.10066965, https://github.com/ShinThant3010/Deep-Learning-based-KG-for-Covid19-Vaccination
Acknowledgment
The authors would like to thank Dr. Chutiporn Anutariya for the fruitful discussion, and her inspirational comments. We would also like to thank the National Institute of Informatics (Tokyo, Japan) for the support of this research. We would like to express all our gratitude to Nicolas Greneche and Prof. Christophe Cerin (the Institut Galilée - LIPN UMR CNRS 7030, UNIVERSITÉ SORBONNE PARIS NORD) for their helpful advice and cooperation in the use of MAGI cloud. This work was also partially supported by JSPS KAKENHI Grant Number JP22K18004.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Thant, S., Racharak, T., Andres, F. (2023). BERT Fine-Tuning the Covid-19 Open Research Dataset for Named Entity Recognition. In: Anutariya, C., Bonsangue, M.M. (eds) Data Science and Artificial Intelligence. DSAI 2023. Communications in Computer and Information Science, vol 1942. Springer, Singapore. https://doi.org/10.1007/978-981-99-7969-1_19
Download citation
DOI: https://doi.org/10.1007/978-981-99-7969-1_19
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-99-7968-4
Online ISBN: 978-981-99-7969-1
eBook Packages: Computer ScienceComputer Science (R0)