Abstract
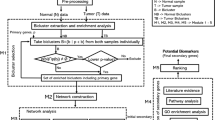
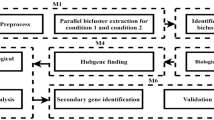
Biclustering has already been established as an effective tool to study gene expression data toward interesting biomarker findings for a given disease. This paper examines the effectiveness of some prominent biclustering algorithms in extracting biclusters of high biological significance toward the identification of interesting biomarkers. We have chosen Esophageal Squamous Cell Carcinoma (ESCC) as a case for our empirical study and our method called BicGenesis could identify eight genes as possible biomarkers for ESCC.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Genecards. https://www.genecards.org/, 31 May 2019
Intogen. https://www.intogen.org/, 31 May 2019
Kemaleren. http://www.kemaleren.com/post/bimax/, 31 May 2019
Malacards. https://malacards.org/, 9 Dec 2018
Yeast saccharomyces cerevisiae cell cycle expression dataset. http://arep.med.harvard.edu/biclustering
Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X et al (2000) Distinct types of diffuse large b-cell lymphoma identified by gene expression profiling. Nature 403(6769):503
Alon U, Barkai N, Notterman DA, Gish K, Ybarra S, Mack D, Levine AJ (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci 96(12):6745–6750
Angiulli F, Cesario E, Pizzuti C (2008) Random walk biclustering for microarray data. Inf Sci 178(6):1479–1497
Bergmann S, Ihmels J, Barkai N (2003) Iterative signature algorithm for the analysis of large-scale gene expression data. Phys Rev E 67(3):031902
Bryan K, Cunningham P, Bolshakova N (2006) Application of simulated annealing to the biclustering of gene expression data. IEEE Trans Inf Technol Biomed 10(3):519–525
Cheng Y, Church GM (2000) Biclustering of expression data. In: ISMB, vol 8, pp 93–103
Chi EC, Allen GI, Baraniuk RG (2017) Convex biclustering. Biometrics 73(1):10–19
Chowdhury HA, Bhattacharyya DK, Kalita JK (2019) (Differential) co-expression analysis of gene expression: a survey of best practices. IEEE/ACM Trans Comput Biol Bioinform 17(4):1154–1173
Clifford RJ, Hu N, Lee MP, Taylor PR (2011) Analysis of gene expression in esophageal squamous cell carcinoma ESCC. NCBI
Coelho GP, de França FO, Von Zuben FJ (2009) Multi-objective biclustering: when non-dominated solutions are not enough. J Math Model Algorithm 8(2):175–202
Dharan S, Nair AS (2009) Biclustering of gene expression data using reactive greedy randomized adaptive search procedure. BMC Bioinform 10(1):S27
Eren K, Deveci M, Küçüktunç O, Çatalyürek ÜV (2012) A comparative analysis of biclustering algorithms for gene expression data. Brief Bioinform 14(3):279–292
Faith JJ, Driscoll ME, Fusaro VA, Cosgrove EJ, Hayete B, Juhn FS, Schneider SJ, Gardner TS (2007) Many microbe microarrays database: uniformly normalized Affymetrix compendia with structured experimental metadata. Nucl Acids Res 36(Suppl 1):D866–D870
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439):531–537
Gu J, Liu JS (2008) Bayesian biclustering of gene expression data. BMC Genomics 9(1):S4
Hartigan JA (1972) Direct clustering of a data matrix. J Am Stat Assoc 67(337):123–129
Hochreiter S, Bodenhofer U, Heusel M, Mayr A, Mitterecker A, Kasim A, Khamiakova T, Van Sanden S, Lin D, Talloen W et al (2010) Fabia: factor analysis for bicluster acquisition. Bioinformatics 26(12):1520–1527
Kluger Y, Basri R, Chang JT, Gerstein M (2003) Spectral biclustering of microarray data: coclustering genes and conditions. Genome Res 13(4):703–716
Lazzeroni L, Owen A (2002) Plaid models for gene expression data. Stat Sinica 61–86
Li G, Ma Q, Tang H, Paterson AH, Xu Y (2009) Qubic: a qualitative biclustering algorithm for analyses of gene expression data. Nucleic Acids Res 37(15):e101–e101
Liu J, Li Z, Hu X, Chen Y (2009) Biclustering of microarray data with mospo based on crowding distance. BMC Bioinform10:S9; BioMed Central (2009)
Lowry DB, Logan TL, Santuari L, Hardtke CS, Richards JH, DeRose-Wilson LJ, McKay JK, Sen S, Juenger TE (2013) Expression quantitative trait locus map** across water availability environments reveals contrasting associations with genomic features in arabidopsis. Plant Cell 25(9):3266–3279
Mandal K, Sarmah R, Bhattacharyya DK (2018) Biomarker identification for cancer disease using biclustering approach: an empirical study. IEEE/ACM Trans Comput Biol Bioinform 16(2):490–509
Mukhopadhyay A, Maulik U, Bandyopadhyay S (2009) A novel coherence measure for discovering scaling biclusters from gene expression data. J Bioinform Comput Biol 7(05):853–868
Murali T, Kasif S (2002) Extracting conserved gene expression motifs from gene expression data. In: Biocomputing 2003. World Scientific, pp 77–88
Network CGA et al (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61
Pontes B, Giráldez R, Aguilar-Ruiz JS (2015) Biclustering on expression data: a review. J Biomed Inform 57:163–180
Pontes Balanza B (2013) Evolutionary biclustering of gene expression data shifting and scaling pattern-based evaluation
Prelić A, Bleuler S, Zimmermann P, Wille A, Bühlmann P, Gruissem W, Hennig L, Thiele L, Zitzler E (2006) A systematic comparison and evaluation of biclustering methods for gene expression data. Bioinformatics 22(9):1122–1129
Tanay A, Sharan R, Shamir R (2002) Discovering statistically significant biclusters in gene expression data. Bioinformatics 18(Suppl 1):S136–S144
Yang J, Wang H, Wang W, Yu PS (2005) An improved biclustering method for analyzing gene expression profiles. Int J Artif Intell Tools 14(05):771–789
Yip KY, Cheung DW, Ng MK (2004) HARP: a practical projected clustering algorithm. IEEE Trans Knowl Data Eng 16(11):1387–1397
Zhang B, Horvath S (2005) A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 4(1)
Zhao L, Zaki MJ (2005) Microcluster: efficient deterministic biclustering of microarray data. IEEE Intell Syst 20(6):40–49
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this paper
Cite this paper
Saikia, M., Bhattacharyya, D.K., Kalita, J.K. (2021). BicGenesis: A Method to Identify ESCC Biomarkers Using the Biclustering Approach. In: Patgiri, R., Bandyopadhyay, S., Balas, V.E. (eds) Proceedings of International Conference on Big Data, Machine Learning and Applications. Lecture Notes in Networks and Systems, vol 180. Springer, Singapore. https://doi.org/10.1007/978-981-33-4788-5_1
Download citation
DOI: https://doi.org/10.1007/978-981-33-4788-5_1
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-33-4787-8
Online ISBN: 978-981-33-4788-5
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)