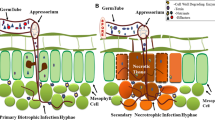
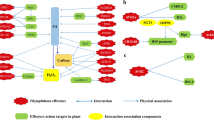
Abstract
Plant-fungal symbioses are of great importance to agriculture. Phytopathogenic fungi, which cause disease in plants as they consume host tissues, are a major threat to global food security. Conversely, endophytes and mycorrhizal fungi, which can engage in mutually beneficial relationships with host plants, are capable of promoting plant health, growth, and development. Such plant-fungal symbioses, whether they be harmful or beneficial to host plants, require complex and continuous molecular cross talk, including the secretion of fungal effector proteins into the plant apoplast and cytoplasm. Fungal effectors are broadly defined as proteins which modulate plant physiology in ways which facilitate fungal colonization and growth within the host plant. These proteins typically share three common features: (1) an amino(N)-terminal signal peptide to allow for secretion, (2) high cysteine content that facilitates structural stability, and (3) small molecular mass (typically ≤30 kDa). However, fungi often possess a large number of genes which fit these criteria—only a subset of which truly act as effector proteins. Progress in identifying effector proteins has been hindered by the fact that most apparently do not share significant sequence similarities with effectors of other genera. This is thought to be an adaptive trait within the context of the co-evolutionary arms race between plant-associated fungi and their host plants, in which plants are continually evolving novel variations in their resistance (R) proteins to allow for improved detection of fungal effector proteins, while fungi diversify their effectors to evade detection. Despite this fact, there are a limited number of domains and motifs which have been found in effector proteins from a variety of plant-associated fungi. Some of these conserved sequence features, such as CFEM and LysM domains, are shared among effector proteins of phytopathogenic and mutualistic fungi, while others may be tailored to the unique lifestyles of individual species or genera. Relying upon the common features and few conserved motifs and domains, in silico tools are often relied upon to mine fungal genomes for candidate effector proteins. Although this process is valuable in curating lists of potential effectors, experimental validation of effector status is still required. To that end, many studies have utilized knockout or overexpression of putative effector protein-coding genes in order to elucidate the importance of putative effectors in the establishment and maintenance of fungal-plant symbioses—a method which is susceptible to issues of gene redundancy. The vast majority of literature on fungal effectors has focused on fungal pathogens of above-ground plant tissue; however this chapter also emphasizes the limited state of understanding on effector proteins from rhizospheric fungi.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Akum FN, Steinbrenner J, Biedenkopf D, Imani J, Kogel KH (2015) The Piriformospora indica effector PIIN_08944 promotes the mutualistic Sebacinalean symbiosis. Front Plant Sci 6:906. https://doi.org/10.3389/fpls.2015.00906
Bailey BA (1995) Purification of a protein from culture filtrates of Fusarium oxysporum that induces ethylene and necrosis in leaves of Erythroxylum coca. Phytopathology 85:1250–1255. https://doi.org/10.1094/Phyto-85-1250
Barelli L, Moonjely S, Behie SW, Bidochka MJ (2016) Fungi with multifunctional lifestyles: endophytic insect pathogenic fungi. Plant Mol Biol 90:657–664. https://doi.org/10.1007/s11103-015-0413-z
Behie SW, Bidochka MJ (2014) Ubiquity of insect-derived nitrogen transfer to plants by endophytic insect-pathogenic fungi: an additional branch of the soil nitrogen cycle. Appl Environ Microbiol 80:1553–1560. https://doi.org/10.1128/AEM.03338-13
Behie SW, Moreira CC, Sementchoukova I, Barelli L, Zelisko PM, Bidochka MJ (2017) Carbon translocation from a plant to an insect-pathogenic endophytic fungus. Nat Commun 8:14,245. https://doi.org/10.1038/ncomms14245
Betsuyaku S, Katou S, Takebayashi Y, Sakakibara H, Nomura N, Fukuda H (2018) Salicylic acid and jasmonic acid pathways are activated in spatially different domains around the infection site during effector-triggered immunity in Arabidopsis thaliana. Plant Cell Physiol 59:8–16. https://doi.org/10.1093/pcp/pcx181
Blümke A, Falter C, Herrfurth C, Sode B, Bode R, Schäfer W et al (2014) Secreted fungal effector lipase releases free fatty acids to inhibit innate immunity-related callose formation during wheat head infection. Plant Physiol 165:346–358. https://doi.org/10.1104/pp.114.236737
Branine M, Bazzicalupo A, Branco S (2019) Biology and applications of endophytic insect-pathogenic fungi. PLoS Pathog 15:e1007831
Brunner-Mendoza C, del Reyes-Montes M, Moonjely S, Bidochka MJ, Toriello C (2019) A review on the genus Metarhizium as an entomopathogenic microbial biocontrol agent with emphasis on its use and utility in Mexico. Biocontrol Sci Tech 29:83–102. https://doi.org/10.1080/09583157.2018.1531111
Castroverde CDM, Nazar RN, Robb J (2016) Verticillium Ave1 effector induces tomato defense gene expression independent of Ve1 protein. Plant Signal Behav 11:e1245254. https://doi.org/10.1080/15592324.2016.1245254
Cen K, Li B, Lu Y, Zhang S, Wang C (2017) Divergent LysM effectors contribute to the virulence of Beauveria bassiana by evasion of insect immune defenses. PLoS Pathog 13:e1006604. https://doi.org/10.1371/journal.ppat.1006604
Crutcher FK, Moran-Diez ME, Ding S, Liu J, Horwitz BA, Mukherjee PK, Kenerley CM (2015) A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol 119:476–486. https://doi.org/10.1016/j.funbio.2015.01.004
De Jonge R, Van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R et al (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955. https://doi.org/10.1126/science.1190859
Djonović S, Pozo MJ, Dangott LJ, Howell CR, Kenerley CM (2006) Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol Plant-Microbe Interact 19:838–853. https://doi.org/10.1094/MPMI-19-0838
Djonović S, Vargas WA, Kolomiets MV, Horndeski M, Wiest A, Kenerley CM (2007) A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol 145:875–889. https://doi.org/10.1104/pp.107.103689
Dölfors F, Holmquist L, Dixelius C, Tzelepis G (2019) A LysM effector protein from the basidiomycete Rhizoctonia solani contributes to virulence through suppression of chitin-triggered immunity. Mol Gen Genomics 294:1211–1218. https://doi.org/10.1007/s00438-019-01573-9
Dou D, Kale SD, Wang X, Chen Y, Wang Q, Wang X et al (2008) Conserved C-terminal motifs required for avirulence and suppression of cell death by Phytophthora sojae effector Avr1b. Plant Cell 20:1118–1133. https://doi.org/10.1105/tpc.107.057067
Dubey MK, Jensen DF, Karlsson M (2014) Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol 14:18. https://doi.org/10.1186/1471-2180-14-18
Dubey M, Vélëz H, Broberg M, Jensen DF, Karlsson M (2020) LysM proteins regulate fungal development and contribute to hyphal protection and biocontrol traits in Clonostachys rosea. Front Microbiol 11:679. https://doi.org/10.3389/fmicb.2020.00679
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC et al (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47:427–432. https://doi.org/10.1093/nar/gky995
Fagan WF, Siemann E, Mitter C, Denno RF, Huberty AF, Woods HA, Elser JJ (2002) Nitrogen in insects: implications for trophic complexity and species diversification. Am Nat 160:784–802. https://doi.org/10.1086/343879
Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ (2012) Emerging fungal threats to animal, plant and ecosystem health. Nature 484:186–194. https://doi.org/10.1038/nature10947
Franceschetti M, Maqbool A, Jiménez-Dalmaroni MJ, Pennington HG, Kamoun S, Banfield MJ (2017) Effectors of filamentous plant pathogens: commonalities amid diversity. Microbiol Mol Biol Rev 81:1–17. https://doi.org/10.1128/mmbr.00066-16
Gaderer R, Lamdan NL, Frischmann A, Sulyok M, Krska R, Horwitz BA, Seidl-Seiboth V (2015) Sm2, a paralog of the Trichoderma cerato-platanin elicitor Sm1, is also highly important for plant protection conferred by the fungal-root interaction of Trichoderma with maize. BMC Microbiol 15:2. https://doi.org/10.1186/s12866-014-0333-0
Gao Q, ** K, Ying SH, Zhang Y, **ao G, Shang Y et al (2011) Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet 7:e1001264. https://doi.org/10.1371/journal.pgen.1001264
Ghareeb H, Drechsler F, Löfke C, Teichmann T, Schirawski J (2015) SUPPRESSOR OF APICAL DOMINANCE1 of Sporisorium reilianum modulates inflorescence branching architecture in maize and arabidopsis. Plant Physiol 169:2789–2804. https://doi.org/10.1104/pp.15.01347
Godfrey D, Böhlenius H, Pedersen C, Zhang Z, Emmersen J, Thordal-Christensen H (2010) Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11:317. https://doi.org/10.1186/1471-2164-11-317
Gorzelak MA, Asay AK, Pickles BJ, Simard SW (2015) Inter-plant communication through mycorrhizal networks mediates complex adaptive behaviour in plant communities. AoB Plants 7:plv050. https://doi.org/10.1093/aobpla/plv022
Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313:903–919. https://doi.org/10.1006/jmbi.2001.5080
Guzmán-Guzmán P, Alemán-Duarte MI, Delaye L, Herrera-Estrella A, Olmedo-Monfil V (2017) Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet 18:1–20. https://doi.org/10.1186/s12863-017-0481-y
Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR et al (2016) Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun 7:11362. https://doi.org/10.1038/ncomms11362
Hassing B, Winter D, Becker Y, Mesarich CH, Eaton CJ, Scott B (2019) Analysis of Epichloë festucae small secreted proteins in the interaction with Lolium perenne. PLoS One 14:e0209463. https://doi.org/10.1371/journal.pone.0209463
Huang A, Lu M, Ling E, Li P, Wang C (2020) A M35 family metalloprotease is required for fungal virulence against insects by inactivating host prophenoloxidases and beyond. Virulence 11:222–237. https://doi.org/10.1080/21505594.2020.1731126
Irieda H, Inoue Y, Mori M, Yamada K, Oshikawa Y, Saitoh H et al (2019) Conserved fungal effector suppresses PAMP-triggered immunity by targeting plant immune kinases. Proc Natl Acad Sci U S A 116:496–505. https://doi.org/10.1073/pnas.1807297116
Jashni MK, Dols IHM, Iida Y, Boeren S, Beenen HG, Mehrabi R et al (2015a) Synergistic action of a metalloprotease and a serine protease from Fusarium oxysporum f. sp. lycopersici cleaves chitin-binding tomato chitinases, reduces their antifungal activity, and enhances fungal virulence. Mol Plant-Microbe Interact 28:996–1008. https://doi.org/10.1094/MPMI-04-15-0074-R
Jashni MK, Mehrabi R, Collemare J, Mesarich CH, de Wit PJGM (2015b) The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front Plant Sci 6:1–7. https://doi.org/10.3389/fpls.2015.00584
Khang CH, Park SY, Lee YH, Valent B, Kang S (2008) Genome organization and evolution of the AVR-pita avirulence gene family in the Magnaporthe grisea species complex. Mol Plant-Microbe Interact 21:658–670. https://doi.org/10.1094/MPMI-21-5-0658
Kloppholz S, Kuhn H, Requena N (2011) A secreted fungal effector of glomus intraradices promotes symbiotic biotrophy. Curr Biol 21:1204–1209. https://doi.org/10.1016/j.cub.2011.06.044
Kombrink A, Rovenich H, Shi-Kunne X, Rojas-Padilla E, van den Berg GCM, Domazakis E et al (2017) Verticillium dahliae LysM effectors differentially contribute to virulence on plant hosts. Mol Plant Pathol 18:596–608. https://doi.org/10.1111/mpp.12520
Kou Y, Tan YH, Ramanujam R, Naqvi NI (2017) Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol 214:330–342. https://doi.org/10.1111/nph.14347
Li S, Peng X, Wang Y, Hua K, **ng F, Zheng Y et al (2019) The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Front Microbiol 10:2228. https://doi.org/10.3389/fmicb.2019.02228
Liu L, Xu L, Jia Q, Pan R, Oelmüller R, Zhang W, Wu C (2019) Arms race: diverse effector proteins with conserved motifs. Plant Signal Behav 14:1557008. https://doi.org/10.1080/15592324.2018.1557008
Lo Presti L, Lanver D, Schweizer G, Tanaka S, Liang L, Tollot M et al (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66:513–545. https://doi.org/10.1146/annurev-arplant-043014-114623
Lyu X, Shen C, Fu Y, **e J, Jiang D, Li G, Cheng J (2016) A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog 12:e1005435. https://doi.org/10.1371/journal.ppat.1005435
Martin F, Aerts A, Ahrén D, Brun A, Danchin EGJ, Duchaussoy F et al (2008) The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature 452:88–92. https://doi.org/10.1038/nature06556
Marton K, Flajsman M, Radisek S, Kosmelj K, Jakse J, Javornik B, Berne S (2018) Comprehensive analysis of Verticillium nonalfalfae in silico secretome uncovers putative effector proteins expressed during hop invasion. PLoS One 13:1–28. https://doi.org/10.1371/journal.pone.0198971
Mei L, Chen M, Shang Y, Tang G, Tao Y, Zeng L et al (2020) Population genomics and evolution of a fungal pathogen after releasing exotic strains to control insect pests for 20 years. ISME J 14:1422–1434. https://doi.org/10.1038/s41396-020-0620-8
Moonjely S, Bidochka MJ (2019) Generalist and specialist Metarhizium insect pathogens retain ancestral ability to colonize plant roots. Fungal Ecol 41:209–217. https://doi.org/10.1016/j.funeco.2019.06.004
Moonjely S, Keyhani NO, Bidochka MJ (2018) Hydrophobins contribute to root colonization and stress responses in the rhizosphere-competent insect pathogenic fungus Beauveria bassiana. Microbiology 164:517–528. https://doi.org/10.1099/mic.0.000644
Moonjely S, Zhang X, Fang W, Bidochka MJ (2019) Metarhizium robertsii ammonium permeases (MepC and Mep2) contribute to rhizoplane colonization and modulates the transfer of insect derived nitrogen to plants. PLoS One 13:e0198971. https://doi.org/10.1371/journal.pone.0223718
Neu E, Debener T (2019) Prediction of the Diplocarpon rosae secretome reveals candidate genes for effectors and virulence factors. Fungal Biol 123:231–239. https://doi.org/10.1016/j.funbio.2018.12.003
Newman MA, Sundelin T, Nielsen JT, Erbs G (2013) MAMP (microbe-associated molecular pattern) triggered immunity in plants. Front Plant Sci 4:139. https://doi.org/10.3389/fpls.2013.00139
Pattemore JA, Hane JK, Williams AH, Wilson BAL, Stodart BJ, Ash GJ (2014) The genome sequence of the biocontrol fungus Metarhizium anisopliae and comparative genomics of Metarhizium species. BMC Genomics 15:660. https://doi.org/10.1186/1471-2164-15-660
Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, De Castro E et al (2015) HAMAP in 2015: updates to the protein family classification and annotation system. Nucleic Acids Res 43:1064–1070. https://doi.org/10.1093/nar/gku1002
Plett JM, Gibon J, Kohler A, Duffy K, Hoegger PJ, Velagapudi R et al (2012) Phylogenetic, genomic organization and expression analysis of hydrophobin genes in the ectomycorrhizal basidiomycete Laccaria bicolor. Fungal Genet Biol 49:199–209. https://doi.org/10.1016/j.fgb.2012.01.002
Plett JM, Daguerre Y, Wittulsky S, Vayssier̀es A, Deveau A, Melton SJ et al (2014) Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci U S A 111:8299–8304. https://doi.org/10.1073/pnas.1322671111
Quarantin A, Hadeler B, Kröger C, Schäfer W, Favaron F, Sella L, Martínez-Rocha AL (2019) Different hydrophobins of fusarium graminearum are involved in hyphal growth, attachment, water-air interface penetration and plant infection. Front Microbiol 10:751. https://doi.org/10.3389/fmicb.2019.00751
Rafiqi M, Jelonek L, Akum NF, Zhang F, Kogel KH (2013) Effector candidates in the secretome of Piriformospora indica, a ubiquitous plant-associated fungus. Front Plant Sci 4:228. https://doi.org/10.3389/fpls.2013.00228
Ramírez-Valdespino CA, Casas-Flores S, Olmedo-Monfil V (2019) Trichoderma as a model to study effector-like molecules. Front Microbiol 10:1030. https://doi.org/10.3389/fmicb.2019.01030
Romero-Contreras YJ, Ramírez-Valdespino CA, Guzmán-Guzmán P, Macías-Segoviano JI, Villagómez-Castro JC, Olmedo-Monfil V (2019) Tal6 From Trichoderma atroviride is a LysM effector involved in mycoparasitism and plant association. Front Microbiol 10:2231. https://doi.org/10.3389/fmicb.2019.02231
Salas-Marina MA, Isordia-Jasso MI, Islas-Osuna MA, Delgado-Sánchez P, Jiménez-Bremont JF, Rodríguez-Kessler M et al (2015) The Epl1 and Sm1 proteins from Trichoderma atroviride and Trichoderma virens differentially modulate systemic disease resistance against different life style pathogens in Solanum lycopersicum. Front Plant Sci 6:77. https://doi.org/10.3389/fpls.2015.00077
Schweizer G, Münch K, Mannhaupt G, Schirawski J, Kahmann R, Dutheil JY (2018) Positively selected effector genes and their contribution to virulence in the smut fungus Sporisorium reilianum. Genome Biol Evol 10:629–645. https://doi.org/10.1093/gbe/evy023
Sȩdzielewska Toro K, Brachmann A (2016) The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genomics 17:101. https://doi.org/10.1186/s12864-016-2422-y
Seidl V, Marchetti M, Schandl R, Allmaier G, Kubicek CP (2006) Epl1, the major secreted protein of Hypocrea atroviridis on glucose, is a member of a strongly conserved protein family comprising plant defense response elicitors. FEBS J 273:4346–4359. https://doi.org/10.1111/j.1742-4658.2006.05435.x
Selin C, de Kievit TR, Belmonte MF, Fernando WGD (2016) Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front Microbiol 7:600. https://doi.org/10.3389/fmicb.2016.00600
Sigrist CJA, De Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A et al (2013) New and continuing developments at PROSITE. Nucleic Acids Res 41:344–347. https://doi.org/10.1093/nar/gks1067
Sonah H, Deshmukh RK, Bélanger RR (2016) Computational prediction of effector proteins in fungi: opportunities and challenges. Front Plant Sci 7:126. https://doi.org/10.3389/fpls.2016.00126
Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF (2007) Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol 16:1701–1711. https://doi.org/10.1111/j.1365-294X.2007.03225.x
Sperschneider J, Dodds PN, Gardiner DM, Singh KB, Taylor JM (2018) Improved prediction of fungal effector proteins from secretomes with EffectorP 2.0. Mol Plant Pathol 19:2094–2110. https://doi.org/10.1111/mpp.12682
Taylor A, Vágány V, Jackson AC, Harrison RJ, Rainoni A, Clarkson JP (2016) Identification of pathogenicity-related genes in Fusarium oxysporum f. sp. cepae. Mol Plant Pathol 17:1032–1347. https://doi.org/10.1111/mpp.12346
Thatcher LF, Gardiner DM, Kazan K, Manners JM (2012) A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol Plant-Microbe Interact 25:180–190. https://doi.org/10.1094/MPMI-08-11-0212
Thatcher LF, Williams AH, Garg G, Buck SAG, Singh KB (2016) Transcriptome analysis of the fungal pathogen Fusarium oxysporum f. sp. medicaginis during colonisation of resistant and susceptible Medicago truncatula hosts identifies differential pathogenicity profiles and novel candidate effectors. BMC Genomics 17:860. https://doi.org/10.1186/s12864-016-3192-2
Tini F, Beccari G, Benfield AH, Gardiner DM, Covarelli L (2020) Role of the XylA gene, encoding a cell wall degrading enzyme, during common wheat, durum wheat and barley colonization by Fusarium graminearum. Fungal Genet Biol 136:103318. https://doi.org/10.1016/j.fgb.2019.103318
Trdá L, Boutrot F, Ciaverie J, Brulé D, Dorey S, Poinssot B (2015) Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline. Front Plant Sci 6:219. https://doi.org/10.3389/fpls.2015.00219
Tsuzuki S, Handa Y, Takeda N, Kawaguchi M (2016) Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol Plant-Microbe Interact 29:277–286. https://doi.org/10.1094/MPMI-10-15-0234-R
Uhse S, Djamei A (2018) Effectors of plant-colonizing fungi and beyond. PLoS Pathog 14:e1006992. https://doi.org/10.1371/journal.ppat.1006992
Van Den Burg HA, Harrison SJ, Joosten MHAJ, Vervoort J, De Wit PJGM (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol Plant-Microbe Interact 19:1420–1430. https://doi.org/10.1094/MPMI-19-1420
Volk H, Marton K, Flajšman M, Radišek S, Tian H, Hein I et al (2019) Chitin-binding protein of Verticillium nonalfalfae disguises fungus from plant chitinases and suppresses chitin-triggered host immunity. Mol Plant-Microbe Interact 32:1378–1390. https://doi.org/10.1094/MPMI-03-19-0079-R
Voß S, Betz R, Heidt S, Corradi N, Requena N (2018) RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front Microbiol 9:2068. https://doi.org/10.3389/fmicb.2018.02068
Wawra S, Fesel P, Widmer H, Timm M, Seibel J, Leson L et al (2016) The fungal-specific β-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat Commun 7:13,188. https://doi.org/10.1038/ncomms13188
van de Wouw AP, Cozijnsen AJ, Hane JK, Brunner PC, McDonald BA, Oliver RP, Howlett BJ (2010) Evolution of linked avirulence effectors in Leptosphaeria maculans is affected by genomic environment and exposure to resistance genes in host plants. PLoS Pathog 6:e1001180. https://doi.org/10.1371/journal.ppat.1001180
Yamamoto N, Wang Y, Lin R, Liang Y, Liu Y, Zhu J et al (2019) Integrative transcriptome analysis discloses the molecular basis of a heterogeneous fungal phytopathogen complex, Rhizoctonia solani AG-1 subgroups. Sci Rep 9:19626. https://doi.org/10.1038/s41598-019-55734-2
Zeng T, Rodriguez-Moreno L, Mansurkhodzaev A, Wang P, van den Berg W, Gasciolli V et al (2020) A lysin motif effector subverts chitin-triggered immunity to facilitate arbuscular mycorrhizal symbiosis. New Phytol 225:448–460. https://doi.org/10.1111/nph.16245
Zhang L, Ni H, Du X, Wang S, Ma XW, Nürnberger T et al (2017) The Verticillium-specific protein VdSCP7 localizes to the plant nucleus and modulates immunity to fungal infections. New Phytol 215:368–381. https://doi.org/10.1111/nph.14537
Zhang Y, Gao Y, Liang Y, Dong Y, Yang X, Qiu D (2019) Verticillium dahliae PevD1, an Alt a 1-like protein, targets cotton PR5-like protein and promotes fungal infection. J Exp Bot 70:613–626. https://doi.org/10.1093/jxb/ery351
Zhu X, Soliman A, Islam MR, Adam LR, Daayf F (2017) Verticillium dahliae’s isochorismatase hydrolase is a virulence factor that contributes to interference with potato’s salicylate and jasmonate defense signaling. Front Plant Sci 8:399. https://doi.org/10.3389/fpls.2017.00399
Zimmermann G (2007) Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci Tech 6:553–596. https://doi.org/10.1080/09583150701309006
Acknowledgments
This research was conducted with the assistance of a Natural Sciences and Engineering Research Council of Canada to MJB (RGPIN-2014-04496).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Stone, L.B.L., Padilla-Guerrero, I.E., Bidochka, M.J. (2022). Fungal Effector Proteins: Molecular Mediators of Fungal Symbionts of Plants. In: Horwitz, B.A., Mukherjee, P.K. (eds) Microbial Cross-talk in the Rhizosphere. Rhizosphere Biology. Springer, Singapore. https://doi.org/10.1007/978-981-16-9507-0_12
Download citation
DOI: https://doi.org/10.1007/978-981-16-9507-0_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-16-9506-3
Online ISBN: 978-981-16-9507-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)