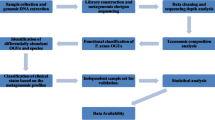
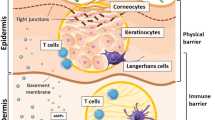
Abstract
The skin is the first barrier of the body and is also a pitch for various microbes. Earlier, research on the skin microbiology was primarily influenced by culture-based techniques. However, now the scenario has changed. Metagenomics has emerged as a more efficient tool for obtaining comprehensive information and understanding of the microbiome of the skin. Metagenomics comprises techniques of high-throughput sequencing, gene-prediction, sequencing of amplicon-based assays, metatranscriptomics and shotgun metagenomics. The sequence data is studied further using statistical as well as comparative analyses. These analyses provide an understanding of the skin microbiome diversity, both functionally and metabolically. Thus, better information of the mechanism of how microbial population of the skin interacts with one other and also with the host could lead to the development of significant clinical treatments as to how we can control those microbial interactions for preventing as well as in curing of various dermatological diseases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Arroyo Mühr LS, Hultin E, Bzhalava D et al (2015) Human papillomavirus type 197 is commonly present in skin tumors. Int J Cancer 136:2546–2555
Barnard E, Shi B, Kang D et al (2016) The balance of metagenomic elements shapes the skin microbiome in acne and health. Sci Rep 6:39491
Byrd AL, Deming C, Cassidy SKB et al (2017) Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med 9:eaal4651
Capone KA, Dowd SE, Stamatas GN, Nikolovski J (2011) Diversity of the human skin microbiome early in life. J Invest Dermatol 131:2026–2032
Chen K, Pachter L (2005) Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput Biol 1:e24–e112
Chen YE, Tsao H (2013) The skin microbiome: current perspectives and future challenges. J Am Acad Dermatol 69:143–155
Cogen AL, Nizet V, Gallo RL (2008) Skin microbiota: a source of disease or defence? Br J Dermatol 158:442–455
Council NR (2007) The new science of metagenomics: revealing the secrets of our microbial planet. National Academies Press, Washington, D.C.
Dawson TL Jr (2007) Malassezia globosa and restricta: breakthrough understanding of the etiology and treatment of dandruff and seborrheic dermatitis through whole-genome analysis. J Investig Dermatol Symp Proc 12:15–19
DeAngelis YM, Gemmer CM, Kaczvinsky JR et al (2005) Three etiologic facets of dandruff and seborrheic dermatitis: Malassezia fungi, sebaceous lipids, and individual sensitivity. J Investig Dermatol Symp Proc 10(3):295–297
Elias PM (2007) The skin barrier as an innate immune element. Semin Immunopathol 29(1):3–14
Fitz-Gibbon S, Tomida S, Chiu B-H et al (2013) Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol 133:2152–2160
Gao Z, Tseng C, Pei Z, Blaser MJ (2007) Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci 104:2927–2932
Grice EA (2015) The intersection of microbiome and host at the skin interface: genomic-and metagenomic-based insights. Genome Res 25:1514–1520
Grice EA, Kong HH, Conlan S et al (2009) Topographical and temporal diversity of the human skin microbiome. Science (80-) 324:1190–1192
Grice EA, Segre JA (2011) The skin microbiome. Nat Rev Microbiol 9:244–253
Grozdev I, Korman N, Tsankov N (2014) Psoriasis as a systemic disease. Clin Dermatol 32:343–350
Guet-Revillet H, Jais J-P, Ungeheuer M-N et al (2017) The microbiological landscape of anaerobic infections in hidradenitis suppurativa: a prospective metagenomic study. Clin Infect Dis 65:282–291
Hannigan GD, Grice EA (2013) Microbial ecology of the skin in the era of metagenomics and molecular microbiology. Cold Spring Harb Perspect Med 3:a015362
Horton JM, Gao Z, Sullivan DM et al (2015) The cutaneous microbiome in outpatients presenting with acute skin abscesses. J Infect Dis 211:1895–1904
Kergourlay G, Taminiau B, Daube G, Vergès M-CC (2015) Metagenomic insights into the dynamics of microbial communities in food. Int J Food Microbiol 213:31–39
Kim M-H, Rho M, Choi J-P et al (2017) A metagenomic analysis provides a culture-independent pathogen detection for atopic dermatitis. Allergy Asthma Immunol Res 9:453–461
Kunin V, Copeland A, Lapidus A et al (2008) A bioinformatician’s guide to metagenomics. Microbiol Mol Biol Rev 72:557–578
Kwon HH, Suh DH (2016) Recent progress in the research about Propionibacterium acnes strain diversity and acne: pathogen or bystander? Int J Dermatol 55:1196–1204
Lau P, Cordey S, Brito F et al (2017) Metagenomics analysis of red blood cell and fresh-frozen plasma units. Transfusion 57:1787–1800
Liu J, Yan R, Zhong Q et al (2015) The diversity and host interactions of Propionibacterium acnes bacteriophages on human skin. ISME J 9:2078–2093
Ma Y, Madupu R, Karaoz U et al (2014) Human papillomavirus community in healthy persons, defined by metagenomics analysis of human microbiome project shotgun sequencing data sets. J Virol 88:4786–4797
Martín R, Miquel S, Langella P, Bermúdez-Humarán LG (2014) The role of metagenomics in understanding the human microbiome in health and disease. Virulence 5:413–423
Mathieu A, Delmont TO, Vogel TM et al (2013) Life on human surfaces: skin metagenomics. PLoS One 8:e65288
Mende DR, Waller AS, Sunagawa S et al (2012) Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS One 7(2):e31386
Ong PY, Ohtake T, Brandt C et al (2002) Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med 347:1151–1160
Proksch E, Brandner JM, Jensen J (2008) The skin: an indispensable barrier. Exp Dermatol 17:1063–1072
Rapp SR, Feldman SR, Exum ML et al (1999) Psoriasis causes as much disability as other major medical diseases. J Am Acad Dermatol 41:401–407
Schuster SC (2008) Next-generation sequencing transforms today’s biology. Nat Methods 5:16–18
Segata N, Waldron L, Ballarini A et al (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814
Segre JA (2006) Epidermal barrier formation and recovery in skin disorders. J Clin Invest 116:1150–1158
Soares RC, Camargo-Penna PH, de Moraes V et al (2016) Dysbiotic bacterial and fungal communities not restricted to clinically affected skin sites in dandruff. Front Cell Infect Microbiol 6:157
Tan H-H (2003) Antibacterial therapy for acne. Am J Clin Dermatol 4:307–314
Tanaka A, Cho O, Saito C et al (2016) Comprehensive pyrosequencing analysis of the bacterial microbiota of the skin of patients with seborrheic dermatitis. Microbiol Immunol 60:521–526
Tett A, Pasolli E, Farina S et al (2017) Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. NPJ Biofilms Microbiomes 3:1–12
Wan T-W, Higuchi W, Khokhlova OE et al (2017) Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS1272 transposition through a novel inverted repeat-replacing mechanism. PLoS One 12:e0187288
White GM (1998) Recent findings in the epidemiologic evidence, classification, and subtypes of acne vulgaris. J Am Acad Dermatol 39:S34–S37
Wilantho A, Deekaew P, Srisuttiyakorn C et al (2017) Diversity of bacterial communities on the facial skin of different age-group Thai males. PeerJ 5:e4084
Wooley JC, Godzik A, Friedberg I (2010) A primer on metagenomics. PLoS Comput Biol 6:e1000667
Zanvit P, Konkel JE, Jiao X et al (2015) Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun 6:1–10
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Singapore Pte Ltd.
About this chapter
Cite this chapter
Sharma, J., Sharma, V., Sharma, I. (2020). The Skin Metagenomes: Insights into Involvement of Microbes in Diseases. In: Chopra, R.S., Chopra, C., Sharma, N.R. (eds) Metagenomics: Techniques, Applications, Challenges and Opportunities. Springer, Singapore. https://doi.org/10.1007/978-981-15-6529-8_12
Download citation
DOI: https://doi.org/10.1007/978-981-15-6529-8_12
Published:
Publisher Name: Springer, Singapore
Print ISBN: 978-981-15-6528-1
Online ISBN: 978-981-15-6529-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)