Abstract
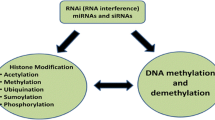
Plant epigenetics has become one of the hottest topics of research not only as a subject of basic research but also as a possible new source of beneficial traits for plant breeding. In addition, epigenetic mechanisms are also crucial to appropriate plant reactions to stress. Given the sessile lifestyle and the late differentiation of the germ line, plants can perceive stresses during vegetative growth and also memorize them, possibly by epigenetic mechanisms. Plants use three systems to initiate and regulate epigenetic gene regulation, like other higher organisms, which include DNA methylation, histone modifications, and RNA interference. New concepts are being evolved to show how these epigenetic components interact and stabilize each other. The role of epigenetic mechanisms in hybrid vigor and epigenetic transgene silencing is also being explored. In this chapter, we have tried to highlight the epigenetic mechanisms that play key roles in plants.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Aina R, Sgorbati S, Santagostino A et al (2004) Specific hypomethylation of DNA is induced by heavy metals in white clover and industrial hemp. Physiol Plant 121(3):472–480
Akimoto K, Katakami H, Kim HJ, Ogawa E, Sano CM, Wada Y, Sano H (2007) Epigenetic inheritance in rice plants. Ann Bot 100(2):205–217
Allen R, Millgate A, Chitty J, Thisleton J, Miller J, Fist A, Gerlach W, Larkin P (2004) RNAi-mediated replacement of morphine with the nonnarcotic alkaloid reticuline in opium poppy. Nat Biotechnol 22:1559–1566
Apel K, Hirt H (2004) Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55:373–399
Berger F (2003) Endosperm: the crossroad of seed development. Curr Opin Plant Biol 6:42–50
Birchler JA, Yao H, Chudalayandi S, Vaimanandveitia RA (2010) Perspective: heterosis. Plant Cell 22:2105–2112
Bird AP (1995) Gene number, noise reduction and biological complexity. Trends Genet 11:94–00
Bird AP (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21
Bird A (2007) Introduction perceptions of epigenetics. Nature 447:396–398
Boscolo PR, Menossi M, Jorge RA (2003) Aluminum-induced oxidative stress in maize. Phytochemistry 62(2):181–189
Boyko A, Kathiria P, Zemp FJ, Yao Y, Pogribny I, Kovalchuk I (2007) Transgenerational changes in the genome stability and methylation in pathogen-infected plants: (virus-induced plant genome instability). Nucleic Acids Res 35(5):1714–1725
Boyko A, Kovalchuk I (2008) Epigenetic control of plant stress response. Environ Mol Mutagen 49(1):61–72
Brettell RI, Dennis ES (1991) Reactivation of a silent Ac following tissue culture is associated with heritable alterations in its methylation pattern. Mol Gen Genet 229(3):365–372
Brink RA (1956) A genetic change associated with the R locus in maize which is directed and potentially reversible. Genetics 41:872–889
Bruce TJA, Matthes MC, Napier JA, Pickett JA (2007) Stressful “memories” of plants: evidence and possible mechanisms. Plant Sci 173:603–608
Cerda S, Weitzman SA (1997) Influence of oxygen radical injury on DNA methylation. Mutat Res 386(2):141–152
Chen F, He G, He H, Chen W, Zhu X, Liang M, Chen L, Deng XW (2010) Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J Integr Plant Biol 52:971–980
Choi CS, Sano H (2007) Abiotic-stress induces demethylation and transcriptional activation of a gene encoding a glycerophosphodiesterase like protein in tobacco plants. Mol Genet Genomics 277:589–600
Choi Y, Gehring M, Johnson L, Hannon M, Harada JJ, Goldberg RB, Jacobsen SE, Fischer RL (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in arabidopsis. Cell 110(1):33–42
Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko JG et al (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461(7262):415–418
De Carvalho F, Gheyson G, Kushnir S, van Montagu M, Inzé D, Castresana C (1992) Suppression of 1,3-glucanase transgene expression in homozygous plants. EMBO J 11:2595–2602
Engler P, Weng A, Storb U (1993) Influence of CpG methylation and target spacing on V(D)J recombination in a transgenic substrate. Mol Cell Biol 13(1):571–577
Fedoroff N (1996) Epigenetic regulation of the maize Spm transposable element. In: Russo VEA, Martienssen RA, Riggs AD (eds) Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, New York, pp 575–592
Feil R, Berger F (2007) Convergent evolution of genomic imprinting in plants and mammals. Trends Genet 23:192–199
Filek M, Keskinen R, Hartikainen H et al (2008) The protective role of selenium in rape seedlings subjected to cadmium stress. J Plant Physiol 165(8):833–844
Fojtova M, Van Houdt H, Depicker A, Kovarik A (2003) Epigenetic switch from posttranscriptional to transcriptional silencing is correlated with promoter hypermethylation. Plant Physiol 133(3):1240–1250
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T (2000) DNA methyltransferases Dnmt1 associates with histone deacetylase activity. Nat Genet 24:88–91
Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278:4035–4040
Gavilano LB, Coleman NP, Burnley LE, Bowman ML, Kalengamaliro NE et al (2006) Genetic engineering of Nicotiana tabacum for reduced nornicotine content. J Agric Food Chem 54:9071–9078
Gehring M, Bubb KL, Henikoff S (2009) Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science 324:1447–1451
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74:481–514
Gong Z, Morales-Ruiz T, Ariza RR et al (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111:803–814
Groszmann M, Greaves IK, Albert N, Fujimoto R, Helliwell CA et al (2011) Epigenetics in plants- vernalisation and hybrid vigour. Biochim Biophys Acta 1809:427–437
Holliday R, Pugh JE (1975) DNA modification mechanisms and gene activity during development. Science 187:226–232
Ingelbrecht I, van Houdt H, van Montagu M, Depicker A (1994) Posttranscriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc Natl Acad Sci U S A 91:10502–10506
Jackson JP, Lindroth AM, Cao X, Jacobsen SE (2002) Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416:556–560
Jacobsen SE, Meyerowitz EM (1997) Hypermethylated SUPERMAN epigenetic alleles in Arabidopsis. Science 277:1100–1103
John MC, Amasino RM (1989) Extensive changes in DNA methylation patterns accompany activation of a silent T-DNA ipt gene in Agrobacterium tumefaciens- transformed plant cells. Mol Cell Biol 9:4298–4303
Johnson L, Cao X, Jacobsen S (2002) Interplay between two epigenetic marks DNA methylation and histone H3 lysine 9 methylation. Curr Biol 12:1360–1367
Jost JP, Siegmann M, Sun L, Leung R (1995) Mechanisms of DNA demethylation in chicken embryos. Purification and properties of a 5-methylcytosine-DNA glycosylase. J Biol Chem 270:9734–9739
Kakutani T (2002) Epi-alleles in plants: inheritance of epigenetic information over generations. Plant Cell Physiol 43:1106–1111
Kalisz S, Purugganan MD (2004) Epialleles via DNA methylation: consequences for plant evolution. Trends Ecol Evol 19:309–314
Kim JM, Ishida TKTJ, Kawashima MM T, Toyoda AMT, Kimura H, Shinozaki K, Seki M (2008) Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 49:1580–1588
Koltunow AM, Grossniklaus U (2003) Apomixis: a developmental perspective. Annu Rev Plant Biol 54:547–574
Kovalchuk I, Kovalchuk O, Kalck V, Boyko V, Filkowski J, Heinlein M, Hohn B (2003) Pathogen-induced systemic plant signal triggers DNA rearrangements. Nature 423:760–762
Kumar SG, Campbell L, Puckhaber L, Stipanovic R, Rathore K (2006) Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proc Natl Acad Sci 103:18054–18059
Labra M, Grassi F, Imazio S et al (2004) Genetic and DNA-methylation changes induced by potassium dichromate in Brassica napus L. Chemosphere 54(8):1049–1058
Lachner M, O’Sullivan RJ, Jenuwein T (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116:2117–2124
Le LQ, Lorenz Y, Scheurer S, Fotisch K, Enrique E et al (2006) Design of tomato fruits with reduced allergenicity by dsRNAi-mediated inhibition of ns-LTP (Lyc e 3) expression. Plant Biotechnol J 4:231–242
Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AHFM (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13:1192–1200
Leonardi A, Damarval C, de Vienne D (1988) Organ-specific variability and inheritance of maize proteins revealed by two-dimensional electrophoresis. Genet Res 52:97–103
Leonardi A, Damerval C, Hebert Y, Gallais A, de Vienne D (1991) Association of protein amount polymorphism (PAP) among maize lines with performances of their hybrids. Theor Appl Genet 82:552–560
Lindbo JA (2012) A historical overview of RNAi in plants. Methods Mol Biol 894:1–16
Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ (2007) Computational and experimental identification of novel human imprinted genes. Genome Res 17(12):1723–1730
Luff B, Pawlowski L, Bender J (1999) An inverted repeat triggers cytosine methylation of identical sequences in Arabidopsis. Mol Cell 3:505–511
Lukens LN, Zhan S (2007) The plant genome’s methylation status and response to stress: implications for plant improvement. Curr Opin Plant Biol 10(3):317–322
Luo M, Liu X, Singh P, Cui Y, Zimmerli L, Wu K (2012) Chromatin modifications and remodeling in plant abiotic stress responses. Biochim Biophys Acta 1819(2):129–136
Mahmoudi T, Verrijzer CP (2001) Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20:3055–3066
Malagnac F, Bartee L, Bender J (2002) An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J 21:6842–6852
Mann JR (2001) Imprinting in the germline. Stem Cells 19:289–294
Martienssen RA (1996) Epigenetic silencing of Mu transposable elements in maize. In: Russo VEA, Martienssen RA, Riggs AD (eds) Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, New York, pp 593–610
Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J (2007) Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell Res 130:851–862
Matzke MA, Matzke A (1995) How and why do plants inactivate homologous (trans)genes? Plant Physiol 107(3):679–685
Matzke MA, Matzke AJ (2004) Planting the seeds of a new paradigm. PLoS Biol 2:582–586
Matzke M, Primig M, Trnovsky J, Matzke A (1989) Reversible methylation and inactivation of marker genes in sequentially transformed plants. EMBO J 8:643–649
Matzke M, Aufsatz W, Kanno T, Daxinger L, Papp I, Mette MF, Matzke AJ (2004) Genetic analysis of RNA-mediated transcriptional gene silencing. Biochim Biophys Acta 1677(1–3):129–141
McClintock B (1951) Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol 16:13–47
Meaney MJ, Ferguson-Smith AC (2010) Epigenetic regulation of the neural transcriptome: the meaning of the marks. Nat Neurosci 13(11):1313–1318
Melquist S, Luff B, Bender J (1999) Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153:401–413
Morel JB, Mourrain P, Beclin C, Vaucheret H (2000) DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol 10:1591–1594
Murphy SK, Jirtle RL (2003) Imprinting evolution and the price of silence. Bioessays 25:577–588
Nan X, Huck-Hui NG, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A (1998) Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386–389
Napoli C, Lemieux C, Jorgensen R (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible cosuppression of homologous genes in trans. Plant Cell 2:279–289
Narlikar GJ, Fan HY, Kingston RW (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475–487
Neuhuber F, Park YD, Matzke AJ, Matzke MA (1994) Susceptibility of transgene loci to homology-dependent gene silencing. Mol Gen Genet 244(3):230–241
Park Y-D, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJM, Matzke MA (1996) Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J 9:183–194
Paszkowski J, Whitham SA (2001) Gene silencing and DNA methylation processes. Curr Opin Plant Biol 4:123–129
Peschke VM, Phillips RL, Gengenbach BG (1991) Genetic and molecular analysis of tissue-culture-derived Ac elements. Theor Appl Genet 82(2):121–129
Regev A, Lamb MJ, Jablonka E (1998) The role of DNA methylation in invertebrates: developmental regulation or genome defense. Mol Biol Evol 15:880–891
Reyes JC, Grossniklaus U (2003) Diverse functions of Polycomb group proteins during plant development. Semin Cell Dev Biol 14:77–84
Richards EJ (1997) DNA methylation and plant development. Trends Genet 13:319–323
Sha AH, Lin XH, Huang JB, Zhang DP (2005) Analysis of DNA methylation related to rice adult plant resistance to bacterial blight based on methylation-sensitive AFLP (MSAP) analysis. Mol Genet Genomics 273:484–490
Shen H, He H, Li J, Chen W, Wang X et al (2012) Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. Plant Cell 24:875–892
Siritunga D, Sayre RT (2003) Generation of cyanogen-free transgenic cassava. Planta 217(3):367–373
Smith C, Watson C, Bird C, Ray J, Schuch W et al (1990) Expression of a truncated tomato polygalacturonase gene inhibits expression of the endogenous gene in transgenic plants. Mol Gen Genet 224:477–548
Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M (2007) Upregulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 227:245–254
Song R, Messing J (2003) Gene expression of a gene family in maize based on noncollinear haplotypes. Proc Natl Acad Sci U S A 100(15):9055–9060
Soppe WJJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF (2002) DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J 21:6549–6559
Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403:41–45
Sun SL, Zhong JQ, Li SH, Wang XJ (2013) Tissue culture-induced somaclonal variation of decreased pollen viability in torenia (Torenia fournieri Lind). Bot Stud 54:36
Tam PL, Zhou SX, Tan SS (1994) X-chromosome activity of the mouse primordial germ cells revealed by the expression of an X-linked lac Z transgene. Development 120:2925–2932
Tamaru H, Selker EU (2001) A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277–283
Tani E, Polidoros AN, Nianiou-Obeidat I, Tsaftaris AS (2005) DNA methylation patterns are differently affected by planting density in maize inbreds and their hybrids. Maydica 50:19–23
Tariq M, Paszkowski J (2004) DNA and histone methylation in plants. Trends Genet 20:244–251
Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J (2003) Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc Natl Acad Sci U S A 100(15):8823–8827
Tsaftaris AS, Kafka M (1998) Mechanisms of heterosis in crop plants. J Crop Prod 1:95–111
Tsaftaris AS, Polidoros AN (1993) Studying the expression of genes in maize parental inbreds and their heterotic and non-heterotic hybrids. In: Bianchi A, Lupotto E, Motto M (eds) Proceedings of XVI Eucarpia Maize and Sorghum Conference, Bergamo, pp 283–292
Tsaftaris AS, Polidoros AN (2000) DNA methylation and plant breeding. Plant Breed Rev 18:87–176
Tsaftaris AS, Polidoros AN, Koumproglou R, Tani E, Kovacevic NM, Abatzidou E (2005) Epigenetic mechanisms in plants and their implications in plant breeding. In: Tuberosa R, Philips R, Gale M (eds) In the wake of the double helix: from the green revolution to the gene revolution. Avenue Media, Bologna, pp 157–171
Tsuji H, Saika H, Tsutsumi N, Hirai A, Nakazono M (2006) Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol 47:995–1003
Vairapandi M, Duker NJ (1993) Enzymic removal of 5-methylcytosine from DNA by a human DNA-glycosylase. Nucleic Acids Res 21(23):5323–5327
Van Blokland R, van der Geest N, Mol J, Kooter J (1994) Transgene-mediated suppression of chalcone synthase expression in Petunia hybrida results from an increase in RNA turnover. Plant J 6:861–877
Van der Krol A, Mur L, Beld M, Mol JNM, Stuitje AR (1990) Flavonoid genes in petunia: addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell 2:291–299
Van Dijk K, Ding Y, Malkaram S, Riethoven JJ, Liu R, Yang J, Laczko P, Chen H, **a Y, Ladunga I, Avramova Z, Fromm M (2010) Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol 10:238
Vanyushin BF (2005) Enzymatic DNA methylation is an epigenetic control for genetic functions of the cell. Biochemistry (Mosc) 70(5):488–499
Wada Y, Miyamoto K, Kusano T, Sano H (2004) Association between up-regulation of stress-responsive genes and hypomethylation of genomic DNA in tobacco plants. Mol Genomics Genet 271:658–666
Waddington CH (1942) The epigenotype. Endeavour 1:18–20
Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286:481–486
Yang JL, Liu LW, Gong YQ et al (2007) Analysis of genomic DNA methylation level in radish under cadmium stress by methylation sensitive amplified polymorphism technique. J Plant Physiol Mol Biol 33(3):219–222
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13(8):335–340
Zadeh AH, Foster GD (2004) Transgenic resistance to tobacco ringspot virus. Acta Virol 48(3):145–152
Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR (2006) Genome-wide high-resolution map** and functional analysis of DNA methylation in Arabidopsis. Cell 126(6):1189–1201
Zhou DX (2009) Regulatory mechanism of histone epigenetic modifications in plants. Epigenetics 4(1):15–18
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer India
About this chapter
Cite this chapter
Munshi, A., Ahuja, Y.R., Bahadur, B. (2015). Epigenetic Mechanisms in Plants: An Overview. In: Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K. (eds) Plant Biology and Biotechnology. Springer, New Delhi. https://doi.org/10.1007/978-81-322-2283-5_12
Download citation
DOI: https://doi.org/10.1007/978-81-322-2283-5_12
Publisher Name: Springer, New Delhi
Print ISBN: 978-81-322-2282-8
Online ISBN: 978-81-322-2283-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)