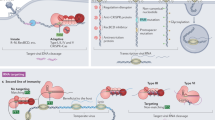
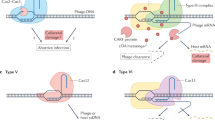
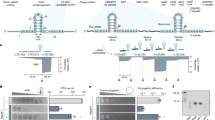
Abstract
CRISPR-Cas is a common adaptive RNA-guided prokaryotic immunity mechanism that limits the spread of mobile genetic elements such as phages and plasmids. A CRISPR-Cas system is composed of two seemingly independent modules. Cas proteins from the adaptation module are responsible for recording prior encounters with mobile genetic elements by incorporating fragments of foreign DNA into CRISPR array. Small protective RNAs generated after CRISPR array transcription are used by the interference module Cas proteins to locate complementary nucleic acids and destroy them. Here, we discuss how the activities and substrate preferences of these two functional modules must be tightly coordinated to provide efficient defence against foreign DNA.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, Severinov K, Regev A, Lander ES, Koonin EV, Zhang F (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353(6299):aaf5573. doi:10.1126/science.aaf5573
Arslan Z, Hermanns V, Wurm R, Wagner R, Pul U (2014) Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucl Acids Res 42(12):7884–7893. doi:10.1093/nar/gku510
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315(5819):1709–1712. doi:10.1126/science.1138140
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321(5891):960–964. doi:10.1126/science.1159689
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340):602–607. doi:10.1038/nature09886
Elmore JR, Sheppard NF, Ramia N, Deighan T, Li H, Terns RM, Terns MP (2016) Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR-Cas system. Genes Dev 30(4):447–459. doi:10.1101/gad.272153.115
Gleditzsch D, Muller-Esparza H, Pausch P, Sharma K, Dwarakanath S, Urlaub H, Bange G, Randau L (2016) Modulating the cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucl Acids Res 44(12):5872–5882. doi:10.1093/nar/gkw469
Goren MG, Doron S, Globus R, Amitai G, Sorek R, Qimron U (2016) Repeat size determination by two molecular rulers in the Type I-E CRISPR array. Cell Rep 16(11):2811–2818. doi:10.1016/j.celrep.2016.08.043
Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP (2009) RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139(5):945–956. doi:10.1016/j.cell.2009.07.040
Hatoum-Aslan A, Samai P, Maniv I, Jiang W, Marraffini LA (2013) A ruler protein in a complex for antiviral defense determines the length of small interfering CRISPR RNAs. J Biol Chem 288(39):27888–27897. doi:10.1074/jbc.M113.499244
Hayes RP, **ao Y, Ding F, van Erp PB, Rajashankar K, Bailey S, Wiedenheft B, Ke A (2016) Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. doi:10.1038/nature16995
Hochstrasser ML, Doudna JA (2015) Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem Sci 40(1):58–66. doi:10.1016/j.tibs.2014.10.007
Kazlauskiene M, Tamulaitis G, Kostiuk G, Venclovas C, Siksnys V (2016) Spatiotemporal control of Type III-A CRISPR-Cas immunity: coupling DNA degradation with the target RNA recognition. Mol Cell 62(2):295–306. doi:10.1016/j.molcel.2016.03.024
Koonin EV, Zhang F (2017) Coupling immunity and programmed cell suicide in prokaryotes: life-or-death choices. BioEssays 39(1):1–9. doi:10.1002/bies.201600186
Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV (2014) Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol 12:36. doi:10.1186/1741-7007-12-36
Kuznedelov K, Mekler V, Lemak S, Tokmina-Lukaszewska M, Datsenko KA, Jain I, Savitskaya E, Mallon J, Shmakov S, Bothner B, Bailey S, Yakunin AF, Severinov K, Semenova E (2016) Altered stoichiometry Escherichia coli cascade complexes with shortened CRISPR RNA spacers are capable of interference and primed adaptation. Nucl Acids Res 44(22):10849–10861. doi:10.1093/nar/gkw914
Luo ML, Jackson RN, Denny SR, Tokmina-Lukaszewska M, Maksimchuk KR, Lin W, Bothner B, Wiedenheft B, Beisel CL (2016) The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucl Acids Res. doi:10.1093/nar/gkw421
Maier LK, Stachler AE, Saunders SJ, Backofen R, Marchfelder A (2015) An active immune defense with a minimal CRISPR (clustered regularly interspaced short palindromic repeats) RNA and without the Cas6 protein. J Biol Chem 290(7):4192–4201. doi:10.1074/jbc.M114.617506
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13(11):722–736. doi:10.1038/nrmicro3569
Marraffini LA, Sontheimer EJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322(5909):1843–1845. doi:10.1126/science.1165771
Marraffini LA, Sontheimer EJ (2010) Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463(7280):568–571. doi:10.1038/nature08703
Moch C, Fromant M, Blanquet S, Plateau P (2016) DNA binding specificities of Escherichia coli Cas1-Cas2 integrase drive its recruitment at the CRISPR locus. Nucl Acids Res. doi:10.1093/nar/gkw1309
Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J (2016) Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353(6299):aad5147. doi:10.1126/science.aad5147
Nunez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA (2015a) Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature 527(7579):535–538. doi:10.1038/nature15760
Nunez JK, Lee AS, Engelman A, Doudna JA (2015b) Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature 519(7542):193–198. doi:10.1038/nature14237
Nunez JK, Bai L, Harrington LB, Hinder TL, Doudna JA (2016) CRISPR immunological memory requires a host factor for specificity. Mol Cell 62(6):824–833. doi:10.1016/j.molcel.2016.04.027
Olovnikov I, Chan K, Sachidanandam R, Newman DK, Aravin AA (2013) Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol Cell 51(5):594–605. doi:10.1016/j.molcel.2013.08.014
Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF (2013) Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell 52(1):124–134. doi:10.1016/j.molcel.2013.08.020
Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA (2015) Co-transcriptional DNA and RNA cleavage during Type III CRISPR-Cas immunity. Cell 161(5):1164–1174. doi:10.1016/j.cell.2015.04.027
Semenova E, Kuznedelov K, Datsenko KA, Boudry PM, Savitskaya EE, Medvedeva S, Beloglazova N, Logacheva M, Yakunin AF, Severinov K (2015) The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR-Cas system. Nucl Acids Res 43(12):6049–6061. doi:10.1093/nar/gkv546
Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, Abudayyeh OO, Gootenberg JS, Makarova KS, Wolf YI, Severinov K, Zhang F, Koonin EV (2017) Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol 15(3):169–182. doi:10.1038/nrmicro.2016.184
Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJ, van der Oost J (2014) DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507(7491):258–261. doi:10.1038/nature12971
Swarts DC, Szczepaniak M, Sheng G, Chandradoss SD, Zhu Y, Timmers EM, Zhang Y, Zhao H, Lou J, Wang Y, Joo C, van der Oost J (2017) Autonomous generation and loading of DNA guides by bacterial argonaute. Mol Cell 65(6):985–998. doi:10.1016/j.molcel.2017.01.033
Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9(8):578–589. doi:10.1038/nrmicro2615
Wang J, Li J, Zhao H, Sheng G, Wang M, Yin M, Wang Y (2015) Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell 163(4):840–853. doi:10.1016/j.cell.2015.10.008
Yoganand KN, Sivathanu R, Nimkar S, Anand B (2017) Asymmetric positioning of Cas1-2 complex and integration host factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucl Acids Res 45(1):367–381. doi:10.1093/nar/gkw1151
Yosef I, Goren MG, Qimron U (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucl Acids Res 40(12):5569–5576. doi:10.1093/nar/gks216
Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, Naismith JH, Spagnolo L, White MF (2012) Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 45(3):303–313. doi:10.1016/j.molcel.2011.12.013
Acknowledgements
This work is supported by an NIH grant GM10407.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Semenova, E., Severinov, K. (2017). Interdependencies Between the Adaptation and Interference Modules Guide Efficient CRISPR-Cas Immunity. In: Pontarotti, P. (eds) Evolutionary Biology: Self/Nonself Evolution, Species and Complex Traits Evolution, Methods and Concepts. Springer, Cham. https://doi.org/10.1007/978-3-319-61569-1_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-61569-1_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-61568-4
Online ISBN: 978-3-319-61569-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)