Abstract
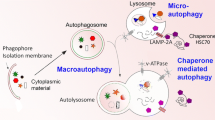
Macroautophagy (autopagy herein) is a cellular stress mechanism characterized by the engulfment of portions of cytoplasm, proteins and organelles in double or multimembrane vesicles. Cargo carried in these autophagic vesicles are then delivered and subsequently degraded by lysosomal/vacuolar systems. Autophagy occurs at low basal levels in all cell types (from yeast to mammals) under non-deprived conditions, performing homeostatic functions. Under conditions leading to cellular stress such as nutrient or growth factor deprivation, autophagy is activated to provide the cell with intracellular building blocks and substrates for energy generation. In addition to the ubiquitin-proteasome system, autophagy is a major degradation pathway for misfolded, mutant or abnormal proteins. Deregulations and abnormalities of autophagy are deleterious for all cell types including neurons. Consequently, autophagy abnormalities are observed in various neuronal diseases. Here, we summarize the basic autophagy machinery and its regulation, and provide a brief summary of the role of autophagy in healthy neurons and in major neurodegenerative diseases.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- ATG:
-
Autophagy related gene
- BCL2:
-
B cell lymphoma 2
- CBZ:
-
Carbamazepine
- CMA:
-
Chaperone mediated autophagy
- DETOR DEP:
-
Domain containing mTOR interacting protein
- FAK:
-
Focal adhesion kinase
- Foxo3:
-
Forkhead box O3
- Hsc70:
-
Heat shock cognate protein 70
- Hvps34:
-
Human vacuolar protein sorting 34
- LAMP2a:
-
Lysosomal associated membrane protein 2 a
- LC3:
-
Microtubule associated protein 1 light chain 3
- Mfns:
-
Mitofusins
- Mtorc1/2:
-
Mammalian Target of Rapamycin Complex 1 and 2
- PEN2:
-
Presenilin enhancer 2
- PI3K:
-
Phosphoinositide 3 kinase
- PINK1:
-
PTEN induced putative kinase1
- PS1/2:
-
Presenilin 1 and 2
- Rag Ras:
-
Related GTP-binding protein
- TSC:
-
Tuberous sclerosis complex
- UPS:
-
Ubiquitin proteasome system
References
Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–75.
Paul S. Dysfunction of the ubiquitin-proteasome system in multiple disease conditions: therapeutic approaches. Bioessays. 2008;30:1172–84.
Todde V, Veenhuis M, van der Klei IJ. Autophagy: principles and significance in health and disease. Biochim Biophys Acta. 2009;1792:3–13.
Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J Cell Biol. 1962;12:198–202.
Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–47.
Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007;104:19500–05.
Curt A, Zhang J, Minnerly J, Jia K. Intestinal autophagy activity is essential for host defense against Salmonella typhimurium infection in Caenorhabditis elegans. Dev Comp Immunol. 2014;45:214–8.
Chargui A, El May MV. Autophagy mediates neutrophil responses to bacterial infection. APMIS. 2014;122:1047–58.
Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22.
Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7.
Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–67.
Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–93.
Ravikumar B, Moreau K, Rubinsztein DC. Plasma membrane helps autophagosomes grow. Autophagy. 2010;6:1184–6.
Wong PM, Puente C, Ganley IG, Jiang XJ. The ULK1 complex sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–37.
Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, et al. (2010) Lysosomal proteolysis and autophagy require Presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell 141: 1146–U1191.
Lee EJ, Tournier C. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011;7:689–95.
Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–502.
Hara T, Takamura A, Kishi C, Iemura S, Natsume T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510.
Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003.
Chan EY, Longatti A, McKnight NC, Tooze SA. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol. 2009;29:157–71.
Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–8.
Foster FM, Traer CJ, Abraham SM, Fry MJ. The phosphoinositide (PI) 3-kinase family. J Cell Sci. 2003;116:3037–40.
Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701.
Panaretou C, Domin J, Cockcroft S, Waterfield MD. Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase. Substrate presentation by phosphatidylinositol transfer protein to the p150.Ptdins 3-kinase complex. J Biol Chem. 1997;272:2477–85.
Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–62.
Russell RC, Tian Y, Yuan H, Park HW, Chang YY, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50.
Proikas-Cezanne T, Codogno P. Beclin 1 or not Beclin 1. Autophag. 2011;7:671–2.
Mauthe M, Jacob A, Freiberger S, Hentschel K, Stierhof YD, et al. Resveratrol-mediated autophagy requires WIPI-1 regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy. 2011;7:1448–61.
Bakula D, Takacs Z, Proikas-Cezanne T. WIPI beta-propellers in autophagy-related diseases and longevity. Biochem Soc Trans. 2013;41:962–7.
Young ARJ, Chan EYW, Hu XW, Koch R, Crawshaw SG, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–900.
Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal. 2011;14:2201–14.
Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012;23:896–909.
Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–75.
Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. Embo J. 2012;31:4304–17.
Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000;151:263–76.
Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92.
Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–96.
Mizushima N, Yoshimori T, Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2003;35:553–61.
Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–12.
Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–18.
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–28.
Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68.
Kimura S, Noda T, Yoshimori T. Dynein-dependent movement of autophagosomes mediates efficient encounters with lysosomes. Cell Struct Funct. 2008;33:109–22.
Anderson RG, Falck JR, Goldstein JL, Brown MS. Visualization of acidic organelles in intact cells by electron microscopy. Proc Natl Acad Sci U S A. 1984;81:4838–42.
Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30.
Scarffe LA, Stevens DA, Dawson VL, Dawson TM. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–24.
Mijaljica D, Prescott M, Devenish RJ. Microautophagy in mammalian cells: revisiting a 40-year-old conundrum. Autophagy. 2011;7:673–82.
Dubouloz F, Deloche O, Wanke V, Cameroni E, De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol Cell. 2005;19:15–26.
Wada Y, Sun-Wada GH, Kawamura N. Microautophagy in the visceral endoderm is essential for mouse early development. Autophagy. 2013;9:252–4.
Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491–9.
Chiang HL, Dice JF. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;263:6797–805.
Salvador N, Aguado C, Horst M, Knecht E. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state. J Biol Chem. 2000;275:27447–56.
Terlecky SR, Dice JF. Polypeptide import and degradation by isolated lysosomes. J Biol Chem. 1993;268:23490–5.
Kaushik S, Massey AC, Mizushima N, Cuervo AM. Constitutive activation of chaperone-mediated autophagy in cells with impaired macroautophagy. Mol Biol Cell. 2008;19:2179–92.
Loos B, Engelbrecht AM, Lockshin RA, Klionsky DJ, Zakeri Z. The variability of autophagy and cell death susceptibility Unanswered questions. Autophagy. 2013;9:1270–85.
Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–34.
Kaushik S, Bandyopadhyay U, Sridhar S, Kiffin R, Martinez-Vicente M, et al. Chaperone-mediated autophagy at a glance. J Cell Sci. 2011;124:495–9.
Fujiwara Y, Furuta A, Kikuchi H, Aizawa S, Hatanaka Y, et al. Discovery of a novel type of autophagy targeting RNA. Autophagy. 2013;9:403–9.
Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–93.
Tato I, Bartrons R, Ventura F, Rosa JL. Amino Acids Activate Mammalian Target of Rapamycin Complex 2 (mTORC2) via PI3K/Akt Signaling. J Biol Chem. 2011;286:6128–42.
Nyfeler B, Bergman P, Triantafellow E, Wilson CJ, Zhu YY, et al. Relieving Autophagy and 4EBP1 from Rapamycin Resistance. Mol Cell Biol. 2011;31:2867–76.
Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–30.
Phung TL, Ziv K, Dabydeen D, Eyiah-Mensch G, Riveros M, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. 2006;10:159–70.
Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol. 2014;15:155–62.
Maiese K, Chong ZZ, Shang YC, Wang S. mTOR: on target for novel therapeutic strategies in the nervous system. Trends Mol Med. 2013;19:51–60.
Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7:e38.
Thoreen CC. Many roads from mTOR to eIF4F. Biochem Soc Trans. 2013;41:913–6.
Zullo AJ, Jurcic Smith KL, Lee S. Mammalian target of Rapamycin inhibition and mycobacterial survival are uncoupled in murine macrophages. BMC Biochem. 2014;15:4.
Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, et al. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303.
Oshiro N, Rapley J, Avruch J. Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging. J Biol Chem. 2014;289:2658–74.
Huang JX, Dibble CC, Matsuzaki M, Manning BD. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–15.
Cai D, Wang Y, Ottmann OG, Barth PJ, Neubauer A, et al. FLT3-ITD-, but not BCR/ABL-transformed cells require concurrent Akt/mTor blockage to undergo apoptosis after histone deacetylase inhibitor treatment. Blood. 2006;107:2094–7.
Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–34.
Salido GM, Sage SO, Rosado JA. TRPC channels and store-operated Ca(2+) entry. Biochem Biophys Acta. 2009;1793:223–30.
Russo R, Berliocchi L, Adornetto A, Varano GP, Cavaliere F, et al. Calpain-mediated cleavage of Beclin-1 and autophagy deregulation following retinal ischemic injury in vivo. Cell Death Dis. 2011;2:e144.
Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–11.
Criollo A, Vicencio JM, Tasdemir E, Maiuri MC, Lavandero S, et al. The inositol trisphosphate receptor in the control of autophagy. Autophagy. 2007;3:350–3.
Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71.
Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–46.
Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, et al. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–68.
Guo K, Searfoss G, Krolikowski D, Pagnoni M, Franks C, et al. Hypoxia induces the expression of the pro-apoptotic gene BNIP3. Cell Death Differ. 2001;8:367–76.
Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009;29:2570–81.
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9.
Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy. 2012;8:858–60.
Rixon HW, Brown G, Murray JT, Sugrue RJ. The respiratory syncytial virus small hydrophobic protein is phosphorylated via a mitogen-activated protein kinase p38-dependent tyrosine kinase activity during virus infection. J Gen Virol. 2005;86:375–84.
Shen W, Ganetzky B. Autophagy promotes synapse development in Drosophila. J Cell Biol. 2009;187:71–9.
Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26:1711–20.
He C, Wei Y, Sun K, Li B, Dong X, et al. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013;154:1085–99.
Hernandez D, Torres CA, Setlik W, Cebrian C, Mosharov EV, et al. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 2012;74:277–84.
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4.
McIntire SL, Garriga G, White J, Jacobson D, Horvitz HR. Genes necessary for directed axonal elongation or fasciculation in C. elegans. Neuron. 1992;8:307–22.
Liang CC, Wang C, Peng X, Gan B, Guan JL. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J Biol Chem. 2010;285:3499–509.
Ban BK, Jun MH, Ryu HH, Jang DJ, Ahmad ST, et al. Autophagy negatively regulates early axon growth in cortical neurons. Mol Cell Biol. 2013;33:3907–19.
Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006;26:8057–68.
Tekirdag KA. Alteration in autophagic-lysosomal potential during ageing and neurological diseases: the microRNA perspective. Curr Pathobiol Rep. 2013;1:247–61.
Citron M, Oltersdorf T, Haass C, McConlogue L, Hung AY, et al. Mutation of the beta-amyloid precursor protein in familial Alzheimer’s disease increases beta-protein production. Nature. 1992;360:672–4.
Scheuner D, Eckman C, Jensen M, Song X, Citron M, et al. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2:864–70.
Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–91.
Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–21.
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118:2190–9.
Li W, Tang Y, Fan Z, Meng Y, Yang G, et al. Autophagy is involved in oligodendroglial precursor-mediated clearance of amyloid peptide. Mol Neurodegener. 2013;8:27.
Jiang T, Yu JT, Zhu XC, Tan MS, Wang HF, et al. Temsirolimus promotes autophagic clearance of amyloid-beta and provides protective effects in cellular and animal models of Alzheimer’s disease. Pharmacol Res. 2014;81:54–63.
Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, et al. Macroautophagy-a novel Beta-amyloid peptide-generating pathway activated in Alzheimer’s disease. J Cell Biol. 2005;171:87–98.
Dauer W, Przedborski S. Parkinson’s disease: mechanisms and models. Neuron. 2003;39:889–909.
Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson’s disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–11.
Bellani S, Sousa VL, Ronzitti G, Valtorta F, Meldolesi J, et al. The regulation of synaptic function by alpha-synuclein. Commun Integr Biol. 2010;3:106–9.
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–13.
Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–5.
Spencer B, Potkar R, Trejo M, Rockenstein E, Patrick C, et al. Beclin 1 gene transfer activates autophagy and ameliorates the neurodegenerative pathology in alpha-synuclein models of Parkinson’s and Lewy body diseases. J Neurosci. 2009;29:13578–88.
Narendra DP, ** SM, Tanaka A, Suen DF, Gautier CA, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298.
Vives-Bauza C, Tocilescu M, Devries RL, Alessi DM, Jackson-Lewis V, et al. Control of mitochondrial integrity in Parkinson’s disease. Prog Brain Res. 2010;183:99–113.
Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14.
West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–7.
Hatano T, Kubo S, Imai S, Maeda M, Ishikawa K, et al. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet. 2007;16:678–90.
Brice A. Genetics of Parkinson’s disease: LRRK2 on the rise. Brain. 2005;128:2760–2.
Cherra SJ, 3rd, Steer E, Gusdon AM, Kiselyov K, Chu CT. Mutant LRRK2 elicits calcium imbalance and depletion of dendritic mitochondria in neurons. Am J Pathol. 2013;182:474–84.
Thomas KJ, McCoy MK, Blackinton J, Beilina A, van der Brug M, et al. DJ-1 acts in parallel to the PINK1/parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50.
Dagda RK, Das Banerjee T, Janda E. How Parkinsonian toxins dysregulate the autophagy machinery. Int J Mol Sci. 2013;14:22163–89.
Yoo MS, Kawamata H, Kim DJ, Chun HS, Son JH. Experimental strategy to identify genes susceptible to oxidative stress in nigral dopaminergic neurons. Neurochem Res. 2004;29:1223–34.
Choi KC, Kim SH, Ha JY, Kim ST, Son JH. A novel mTOR activating protein protects dopamine neurons against oxidative stress by repressing autophagy related cell death. J Neurochem. 2010;112:366–76.
Ha JY, Kim JS, Kang YH, Bok E, Kim YS, et al. Tnfaip8 l1/Oxi-beta binds to FBXW5, increasing autophagy through activation of TSC2 in a Parkinson’s disease model. J Neurochem. 2014;129:527–38.
Holmberg CI, Staniszewski KE, Mensah KN, Matouschek A, Morimoto RI. Inefficient degradation of truncated polyglutamine proteins by the proteasome. EMBO J. 2004;23:4307–18.
Sarkar S, Rubinsztein DC. Huntington’s disease: degradation of mutant huntingtin by autophagy. FEBS J. 2008;275:4263–70.
Bursch W, Ellinger A. Autophagy-a basic mechanism and a potential role for neurodegeneration. Folia Neuropathol. 2005;43:297–310.
Santos RX, Cardoso S, Correia S, Carvalho C, Santos MS, et al. Targeting autophagy in the brain: a promising approach? Cent Nerv Syst Agents Med Chem. 2010;10:158–68.
Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–95.
Martin DD, Heit RJ, Yap MC, Davidson MW, Hayden MR, et al. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of Huntingtin. Hum Mol Genet. 2014;23:3166–79.
Krainc D. Clearance of mutant proteins as a therapeutic target in neurodegenerative diseases. Arch Neurol. 2010;67:388–92.
Rubinsztein DC, Ravikumar B, Acevedo-Arozena A, Imarisio S, O’Kane CJ, et al. Dyneins, autophagy, aggregation and neurodegeneration. Autophagy. 2005;1:177–8.
Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42.
Acknowledgements
This work was supported by The Scientific and Technological Research Council of Turkey (TUBITAK) 1001 Grant Project Number 110T405 and Sabanci University. D.G. is a recipient of the Turkish Academy of Sciences (TUBA) GEBIP Award and the EMBO Strategical Development and Installation Grant (EMBO-SDIG).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Arachiche, A., Gozuacik, D. (2015). Regulation of Autophagy in Health and Disease. In: Fuentes, J. (eds) Toxicity and Autophagy in Neurodegenerative Disorders. Current Topics in Neurotoxicity, vol 9. Springer, Cham. https://doi.org/10.1007/978-3-319-13939-5_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-13939-5_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-13938-8
Online ISBN: 978-3-319-13939-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)