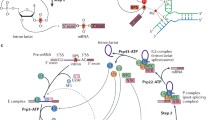
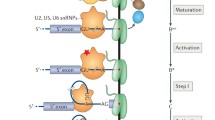
Abstract
At the level of gene architecture, the widespread presence of interrupting sequences in eukaryotic genes serves as a defining difference between eukaryotic organisms and other domains of life. These interrupting sequences, known as introns, must be precisely removed from pre-messenger RNA (pre-mRNA) transcripts. Concomitantly, the coding regions, or exons, are joined together through a nuclear-localized process known as pre-mRNA splicing. A number of splicing factors, both protein and RNA, assemble into a multimegadalton splicing machine known as the spliceosome, which is responsible for identifying the intronic regions and positioning the pre-mRNA substrate in a favorable orientation for the splicing reactions to occur. While the chemical steps of splicing—two sequential transesterification reactions—are identical in all eukaryotes, the gene architecture and splicing apparatus can differ substantially. Here, we review our current understanding of the splicing process with an emphasis on the model organism Saccharomyces cerevisiae. We discuss the key features of introns, along with mechanistic aspects of the splicing cycle, namely spliceosome assembly, catalysis, and spliceosome disassembly. We also highlight recent discoveries supporting the role of kinetic proofreading in ensuring the fidelity of splicing.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abelson J (2013) Toggling in the spliceosome. Nat Struct Mol Biol 20:645–647
Abovich N, Liao XC, Robash M (1994) The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev 8:843–854
Abovich N, Rosbash M (1997) Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell 89:403–412
Arenas JE, Abelson J (1997) Prp43: an RNA helicase-like factor involved in spliceosome disassembly. PNAS 94:11798–11802
Ares M Jr, Grate L, Pauling MH (1999) A handful of intron-containing genes produces the lion’s share of yeast mRNA. RNA 5:113–1139
Bartels C, Urlaub H, Lührmann R et al (2003) Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem 278:28324–28334
Bell M, Schreiner S, Damainov A et al (2002) p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21:2724–2735
Bellare P, Small EC, Huang X et al (2008) A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol 15:444–451
Bon E, Casaregola S, Blandin G et al (2003) Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. NAR 31:1121–1135
Brenner TJ, Guthrie C (2005) Genetic analysis reveals a role for the C terminus of the Sacchaomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 170:1063–1080
Brenner TJ, Guthrie C (2006) Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. RNA 12:862–871
Brody E, Abelson J (1985) The “Spliceosome”: Yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science 228:963–967
Brow DA, Guthrie C (1988) Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334:213–218
Brow DA, Guthrie C (1989) Splicing a spliceosomal RNA. Nature 337:14–15
Brow DA (2002) Allosteric cascade of spliceosomal activation. Annu Rev Genet 36:333–360
Brys A, Schwer B (1996) Requirement for Slu7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and 3′ splice site. RNA 2:707–717
BurgessSM Guthrie C (1993) A mechanism to enhance mRNA splicing fidelity: the RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73:1377–1391
Burset M, Seledtov IA, Solovyev VV (2000) Analysis of canonical and non-canonicle splice sites in mammalian genomes. NAR 28:4364–4375
Cech T (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44:207–210
Chan SP, Kao DI, Tsai WY et al (2003) The Prp19p-associated complex in splicesome activation. Science 302:279–282
Chan SP, Cheng SC (2005) The Prp19-associated complex is required for specifying interactions of U5 and U6 with the pre-mRNA during spliceosome activation. J Biol Chem 280:31190–31199
Chen HC, Cheng SC (2012) Functional roles of protein splicing factors. Biosci Rep 32:345–359
Chen HC, Tseng CK, Tsai RT et al (2013) Link of NTR-mediated spliceosome disassembly with DEAH-box ATPases Prp2, Prp16, and Prp22. Mol Cell Biol 33:514–525
Chen JY, Stands L, Staley JP et al (2001) Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol Cell 7:227–232
Cheng SC, Abelson J (1987) Spliceosome assembly in yeast. Genes Dev 1:1014–1027
Chua K, Reed R (1999) The RNA splicing factor hSlu7 is required for correct 3′ splice site choice. Nature 402:207–210
Collemare J, van der Burget A, de Wit PJ (2013) At the origin of spliceosomal introns: is multiplication of intron-like elements the main mechanism of intron gain in fungi? Commun. Integr. Biol. 6(2):e23147
Company M, Arenas J, Abelson J (1991) Requirement of the RNA helicase-like Prp22 for release of messenger RNA from spliceosomes. Nature 349:487–493
Coulombe-Huntington J, Majewski J (2007) Characterization of intron loss events in mammals. Genome Res 17:23–32
Crawford DJ, Hoskins AA, Friedman LJ et al (2013) Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. PNAS 110:6783–6788
Deutsch M, Long M (1999) Intron-exon structures of eukaryotic model organisms. NAR 27:3219–3228
Dlakic M, Mushegian A (2011) Prp8, the pivotal protein of the spliceosomal catalytic center, evolved from a retroelement-encoded reverse transcriptase. RNA 17:799–808
Dix I, Russell CS, O’Keefe RT et al (1998) Protein–RNA interactions in the U5 snRNP of Saccharomyces ceravisiae. RNA 4:1675–1686
Fabrizio P, Abelson J (1992) Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. NAR. 20:3659–3664
Fabrizio P, Laggerbauer B, Lauber J et al (1997) An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO 16:4092–4106
Fabrizio P, Dannenberg J, Dube P et al (2009) The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36:593–608
Farlow A, Meduri E, Schlötterer C (2011) DNA double-strand break repair and the evolution of intron density. Trends Genet 27:1–6
Fica SM, Tuttle N, Novak T et al (2013) RNA catalyses nuclear pre-mRNA splicing. Nature 503:229–234
Fink GR (1987) Pseudogenes in yeast? Cell 49:5–6
Fortner DM, Troy RG, Brow DA (1994) A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev 8:221–233
Fourmann JB, Schmitzová J, Christian H et al (2013) Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 27:413–428
Fromont-Racine M, Mayes AE, Brunet-Simon A et al (2000) Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17:95–110
Gordon PM, Piccirilli JA (2001) Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol 8:893–898
Gozani O, Feld R, Reed R (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of splicesomal complex A. Genes Dev 10:233–243
Gozani O, Potashkin J, Reed R (1998) A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18:4752–4760
Grabowski PJ, Seiler SR, Sharp PA (1985) A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell 42:345–353
Guthrie C, Patterson B (1988) Spliceosomal snRNAs. Annu Rev Genet 22:387–419
Hahn D, Kudla G, Tollervey D et al (2012) Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev 26:2408–2421
Hilliker AK, Staley JP (2004) Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10:921–928
Hilliker AK, Mefford MA, Staley JP (2007) U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21:821–834
Hopfield JJ (1974) Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. PNAS 71:4135–4139
Hoskins AA, Friedman LJ, Gallagher SS et al (2011) Ordered and dynamic assembly of single spliceosomes. Science 331:1289–1295
Jackson SP, Lossky M, Beggs JD (1988) Cloning of the RNA8 gene of Saccharomyces cerevisiae, detection of the RNA8 protein, and demonstration that it is essential for nuclear pre-mRNA splicing. Mol Cell Biol 8:1067–1075
James SA, Turner W, Schwer B (2002) How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068–1077
Jurica MS, Moore MJ (2003) Pre-mRNA splicing: awash in a sea of proteins. Mol Cell 12:5–14
Käufer NF, Potashkin J (2000) Analysis of the splicing machinery in fission yeast: a comparison with budding yeast and mammals. NAR 28:3003–3010
Kistler AL, Guthrie C (2001) Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev 15:42–49
Konarska MM, Grabowski PJ, Padgett RA et al (1985) Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature 313:552–557
Koodathingal P, Novak T, Piccirilli JA et al (2013) The DEAH-box ATPase Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell 39:385–395
Kuhn AN, Li Z, Brow DA (1999) Splicing factor Prp8 governs U4/U6 RNA unwinding during activation of the spliceosome. Mol Cell 3:65–75
Kuhn AN, Reichl EM, Brow DA (2002) Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. PNAS 99:9145–9149
Kwan SS, Brow DA (2005) The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 binding. RNA 11:808–820
Lardelli RM, Thompson JX, Yates JR et al (2010) Release of SF3 from the intron branchpoint activates the first catalytic step of pre-mRNA splicing. RNA 16:516–528
Lee C, Jaladat Y, Mohammadi A et al (2010) Metal binding and substrate positioning by evolutionarily invariant U6 sequences in catalytically active protein-free snRNAs. RNA 16:2226–2238
Legrain P, Séraphin B, Robash M (1985) Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol 8:3755–3760
Li Z, Brow D (1996) A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA 2:879–894
Liu HL, Cheng SC (2012) The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 32:5056–5066
Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71:803–817
Maeder C, Kutach AK, Guthrie C (2009) ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16:42–48
Maschhloff K, Padgett R (1993) The stereochemical course of the first step of pre-mRNA splicing. NAR. 21:5456–5462
Mayas RM, Maita H, Staley JP (2006) Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol 13:482–490
Mayes AE, Verdone L, Legrain P et al (1999) Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18:4321–4331
McGrail JC, Krause A, O’Keefe RT (2009) The RNA binding protein Cwc2 interacts directly with the U6 snRNA to link the nineteen complex to the spliceosome during pre-mRNA splicing. NAR 37:4205–4217
Mefford MA, Staley JP (2009) Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 15:1386–1397
Mekouar M, Blanc-Lenfle I, Ozanne C et al (2010) Detection and analysis of alternative splicing in Yarrowia lipolytica reveal structural constraints facilitating nonsense-mediated decay of intron-retaining transcripts. Genome Biol 11:R65. doi:10.1186/gb-2010-11-6-r65
Mitrovich QM, Guthrie C (2007) Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA 13:2066–2080
Moore MJ, Sharp PA (1993) Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365:364–368
Mozaffari-Jovin S, Santos KF, Hsiao HH et al (2012) The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev 26:2422–2434
Mozaffari-Jovin S, Wandersleben T, Santos KF et al (2013) Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 341:80–84
Neuvéglise C, Marck C, Gaillardin C (2011) The intronome of budding yeasts. C R Biol 34:662–670
Newman AJ, Teigelkamp S, Beggs JD (1995) snRNA interactions at 5′ and 3′ splice sites monitored by photoactivated crosslinking in yeast spliceosomes. RNA 1:968–980
Nielsen KH, Staley JP (2012) Spliceosome activation: U4 is the path, stem I is the goal, and Prp8 is the keeper. Let’s cheer for the ATPase Brr2! Genes Dev 26:2461–2467
Ninio J (1975) Kinetic amplification of enzyme discrimination. Biochimie 57:587–595
O’Day CL, Dalbadie-McFarland G, Abelson J (1996) The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem 271:33261–33267
Ohi MD, Vander Kooi CW, Rosenberg JA et al (2003) Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol 10:250–255
Ohrt T, Odenwälder P, Dannenberg J et al (2013) Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA 19:902–915
Oubridge C, Ito N, Evans PR et al (1994) Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372:432–438
Padgett RA, Konarska MM et al (1984) Lariat RNAs as intermediates and products in the splicing of messenger RNA precursors. Science 225:898–903
Parker R, Siliciano PG, Guthrie C (1987) Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49:229–239
Patterson B, Guthrie C (1987) An essential yeast snRNA with a U5-like domain is required for splicing in vivo. Cell 49:613–624
Peebles CL, Zhang M, Perlman PS et al (1995) Catalytically critical nucleotides in domain 5 of a group II intron. PNAS 92:4422–4426
Pena V, Rozov A, Fabrizio P et al (2008) Structure and function of an RNAse H domain at the heart of the splicesome. EMBO J 27:2929–2940
Puig O, Gottschalk A, Fabrizio P et al (1999) Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev 13:569–580
Puig O, Bragado-Nilsson E, Koski T et al (2007) The U1 snRNP-associated factor Luc7p affects 5′ splice site selection in yeast and human. NAR 35:5874–5885
Rader SD, Guthrie C (2002) A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8:1378–1392
Raghunathan PL, Guthrie C (1998a) A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 29:857–860
Raghunathan PL, Guthrie C (1998b) RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8:847–855
Rhode BM, Hartmuth K, Westhof E et al (2006) Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J 25:2475–2486
Rymond BC, Rosbash M (1985) Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature 317:735–737
Sakharkar K, Chow V, Kangueane P (2004) Distribution of exons and introns in the human genome. In Silico Biol 4:387–393
Schellenberg MJ, Wu T, Ritchie DB et al (2013) A conformational switch in Prp8 mediates metal ion coordination that promotes pre-mRNA exon ligation. Nat Struct Mol Biol 20:728–724
Schwer B, Guthrie C (1991) Prp16 is an RNA-dependent ATPase that interacts transiently with the spliceisome. Nature 349:494–499
Schwer B, Guthrie C (1992) A conformational rearrangement in the spliceosome is dependent on Prp16 and ATP hydrolysis. J EMBO 11:5033–5039
Schwer B, Gross CH (1998) Prp22, a DExD-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J 17:2086–2094
Schwer B (2008) A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30:743–754
Schwer B, Chang J, Shuman S (2013) Structure-function analysis of the 5′ end of yeast U1 snRNA highlights genetic interactions with the Msl5∙Mud2 branchpoint-binding complex and other spliceosome assembly factors. NAR 41:7485–7500
Semlow DR, Staley JP (2012) Staying on message: Ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci 37:263–273
Shannon KW, Guthrie C (1991) Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev 5:773–785
Shuster EO, Guthrie C (1988) Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell 55:41–48
Shuster EO, Guthrie C (1990) Human U2 snRNA can function in pre-mRNA splicing in yeast. Nature 345:270–273
Sigel RK, Vaidya A, Pyle AM (2000) Metal ion binding sites in a group II intron core. Nat Struct Biol 7:1111–1116
Siliciano PG, Guthrie C (1988) 5′ splice site selection in yeast: genetic alterations in base-pairing with U1 reveal additional requirements. Genes Dev 2:1258–1267
Small EC, Leggett SR, Winans AA et al (2006) The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23:389–399
Song EJ, Werner SL, Neubauer J et al (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev 24:1434–1447
Sontheimer EJ, Steitz JA (1993) The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262:1989–1996
Soochin C, Suk-Won J, Cohen A et al (2004) A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res 14:1207–1220
Staley JP, Guthrie C (1999) An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3:55–64
Steitz TA, Steitz JA (1993) A general two-metal-ion mechanism for catalytic RNA. PNAS 90:6498–6502
Stevens SW, Barta I, Ge HY et al (2001) Biochemical and genetic analyses of the U5, U6, and U4/U6∙U5 small nucler ribonucleoproteins from Saccharomyces cerevisiea. RNA 7:1543–1553
Stevens SW, Ryan DE, Ge HY et al (2002) Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell 9:31–44
S**ola M, Grate L, Haussler D et al (1999) Genome-wide bioinformatics and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221–234
Surowy CS, van Santen VL, Scheib-Wixted SM et al (1989) Direct, sequence specific binding of the human U1-70 k ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol Cell Biol 9:4179–4186
Tanaka N, Aronova A, Schwer B (2007) Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21:2312–2325
Tsai RT, Fu RH, Yeh FL et al (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19:2991–3003
Tsai RT, Tseng CK, Lee PJ et al (2007) Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. J Mol Cell Biol 27:8027–8037
Tseng CK, Liu HL, Cheng SC (2011) DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17:145–154
Valadkhan S, Manley J (2001) Splicing-related catalysis by protein-free snRNAs. Nature 413:701–707
Van der Veen R, Arnberg A, van der Horst G et al (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44:225–234
Venter JC, Adams MD, Myers EW et al (2001) The sequence of the human genome. Science 291:1304–1351
Vidovic I, Nottrott S, Hartmuth K et al (2000) Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6:1331–1342
Vinogradov AE (2001) Intron length and codon usage. J Mol Evol 52:2–5
Wang Q, Zhang L, Lynn B et al (2008) A BBP-Mud2p heterodimer mediates branchpoint recognition and influences splicing substrate abundance in budding yeast. NAR 36:2787–2798
Warkocki Z, Odenwälder P, Schmitzová J et al (2009) Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16:1237–1243
Weber S, Aebi M (1988) In vitro splicing of mRNA precursors: 5′ cleavage site can be predicted from the interaction between the 5′ splice site region and the 5′ terminus of U1 snRNA. NAR 16:471–486
Wiest DK, O’Day CL, Abelson J (1996) In vitro studies of the Prp9∙Prp11∙Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem 271:33268–33276
Wood V, Gwilliam R, Rajandream MA et al (2002) The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880
Wu S, Romfo CM, Nilsen TW et al (1999) Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832–835
Xu D, Nouraini S, Field D et al (1996) An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature 381:709–713
Xu YZ, Query CC (2007) Competition between the ATPase Prp5 and branch region U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell 28:838–849
Yang F, Wang X, Zhang ZM et al (2013) Splicing proofreading at 5′ splice sites by ATPase Prp28p. NAR 41:4660–4670
Yean SL, Wuenschell G, Termini J et al (2000) Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408:881–884
Zhu T, Niu DK (2013) Mechanisms of intron loss and gain in the fission yeast Schizosaccharomyces. PLoS ONE 8:e61683
Acknowledgments
This work was supported by NSERC Discovery Grant 298521 to SDR and an NSERC PGS award to EAD, as well as by awards from UNBC’s Office of Research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Dunn, E.A., Rader, S.D. (2014). Pre-mRNA Splicing and the Spliceosome: Assembly, Catalysis, and Fidelity. In: Sesma, A., von der Haar, T. (eds) Fungal RNA Biology. Springer, Cham. https://doi.org/10.1007/978-3-319-05687-6_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-05687-6_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-05686-9
Online ISBN: 978-3-319-05687-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)