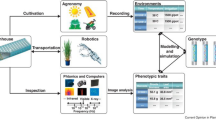
Abstract
The integration of genomics with phenomics leads to efficient breeding and the development of climate-resilient and crop varieties adaptable to the needs of modern breeding. Next-generation high-throughput approaches and plant phenoty** platforms have enabled the efficient, precise, and robust assessment of multiple plant traits in the last two decades. These approaches also mediate the relationship between plant growth and development traits on one hand and reproduction under diverse environmental conditions on the other. Nevertheless, recent high-tech advances develop novel tools with potential solutions to explore large-scale phenoty** data acquisition and processing in the coming years. In this book chapter, we discuss the significant achievement and advancement in high-throughput and phenomics in controlled environmental conditions and its uses for microphenoty**. We also discuss the latest multitudinal genomics research aided with high-throughput phenoty** with plant genetic studies. Finally, we propose few conceptual challenges and provide our future perspectives on bridging the phenotype-genotype-envirotype gap.
Graphical Abstract

Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Al-Tamimi N, Brien C, Oakey H, Berger B, Saade S, Ho YS, Schmöckel SM, Tester M, Negrão S (2016) Salinity tolerance loci revealed in rice using high-throughput non-invasive phenoty**. Nat Commun 7:1–11
Amal TC, Thottathil AT, Veerakumari KP, Rakkiyappan R, Vasanth K (2020) Morphological traits of drought tolerant horse gram germplasm: classification through machine learning. J Sci Food Agric 100:4959–4967
Atkinson JA, Pound MP, Bennett MJ, Wells DM (2019) Uncovering the hidden half of plants using new advances in root phenoty**. Curr Opin Biotechnol 55:1–8
Awlia M, Nigro A, Fajkus J, Schmoeckel SM, Negrão S, Santelia D, Trtílek M, Tester M, Julkowska MM, Panzarová K (2016) High-throughput non-destructive phenoty** of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front in Plant Sci. 7:1414
Bai G, Ge Y, Hussain W, Baenziger PS, Graef G (2016) A multi-sensor system for high throughput field phenoty** in soybean and wheat breeding. Comput Electron Agric 128:181–192
Barone A, Chiusano ML, Ercolano MR, Giuliano G, Grandillo S, Frusciante L (2008) Structural and functional genomics of tomato. Int J Plant Genomics 2008:820274–820274
Bergsträsser S, Fanourakis D, Schmittgen S, Cendrero-Mateo MP, Jansen M, Scharr H, Rascher U (2015) HyperART: non-invasive quantification of leaf traits using hyperspectral absorption-reflectance-transmittance imaging. Plant Methods 11:1–17
Beyer S, Daba S, Tyagi P, Bockelman H, Brown-Guedira G, Mohammadi M (2019) Loci and candidate genes controlling root traits in wheat seedlings—a wheat root GWAS. Funct Integr Genomics 19:91–107
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Bouché N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4:111–117
Bucksch A, Burridge J, York LM, Das A, Nord E, Weitz JS, Lynch JP (2014) Image-based high-throughput field phenoty** of crop roots. Plant Physiol 166:470–486
Burridge J, Jochua CN, Bucksch A, Lynch JP (2016) Legume shovelomics: high—throughput phenoty** of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp, unguiculata) root architecture in the field. Field Crop Res 192:21–32
Busemeyer L, Mentrup D, Möller K, Wunder E, Alheit K, Hahn V, Maurer HP, Reif JC, Würschum T, Müller J (2013) BreedVision—A multi-sensor platform for non-destructive field-based phenoty** in plant breeding. Sensors 13:2830–2847
Bylesjö M, Segura V, Soolanayakanahally RY, Rae AM, Trygg J, Gustafsson P, Jansson S, Street NR (2008) LAMINA: a tool for rapid quantification of leaf size and shape parameters. BMC Plant Biol 8:1–9
Burton AL, Williams M, Lynch JP, Brown KM (2012) RootScan: software for high-throughput analysis of root anatomical traits. Plant and Soil 357(1):189–203
Chimungu JG, Loades KW, Lynch JP (2015) Root anatomical phenes predict root penetration ability and biomechanical properties in maize (Zea mays). J Experi Bot 66(11):3151–3162
Chopin J, Laga H, Huang CY, Heuer S, Miklavcic SJ (2015) Rootanalyzer: a cross-section image analysis tool for automated characterization of root cells and tissues. PloS one 10(9):e0137655
Carpentier S, Costa C, Schurr U, Loreto F, Menesatti P (2019) Plant phenoty** research trends, a science map** approach. Front Plant Sci 9
Casanova JJ, O’Shaughnessy SA, Evett SR, Rush CM (2014) Development of a wireless computer vision instrument to detect biotic stress in wheat. Sensors 14:17753–17769
Chaerle L, Van Der Straeten D (2000) Imaging techniques and the early detection of plant stress. Trends Plant Sci 5:495–501
Chen D, Neumann K, Friedel S, Kilian B, Chen M, Altmann T, Klukas C (2014) Dissecting the phenotypic components of crop plant growth and drought responses based on high-throughput image analysis. Plant Cell 26:4636–4655
Clauw P, Coppens F, De Beuf K, Dhondt S, Van Daele T, Maleux K, Storme V, Clement L, Gonzalez N, Inzé D (2015) Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol 167:800–816
Costa JM, Marques da Silva J, Pinheiro C, Barón M, Mylona P, Centritto M, Haworth M, Loreto F, Uzilday B, Turkan I (2019) Opportunities and limitations of crop phenoty** in southern European countries. Front Plant Sci 10:1125
Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Gozé E, Kilian A (2013) Genome-wide association map** of root traits in a japonica rice panel. PLoS One 8:e78037
Crowell S, Korniliev P, Falcao A, Ismail A, Gregorio G, Mezey J, McCouch S (2016) Genome-wide association and high-resolution phenoty** link Oryza sativa panicle traits to numerous trait-specific QTL clusters. Nat Commun 7:1–14
Davis BD (1949) The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci U S A 35:1
De Bei R, Cozzolino D, Sullivan W, Cynkar W, Fuentes S, Dambergs R, Pech J, Tyerman S (2011) Non-destructive measurement of grapevine water potential using near infrared spectroscopy. Aust J Grape Wine Res 17:62–71
De Diego N, Fürst T, Humplík JF, Ugena L, Podlešáková K, Spíchal L (2017) An automated method for high-throughput screening of Arabidopsis rosette growth in multi-well plates and its validation in stress conditions. Front Plant Sci 8:1702
De Vylder J, Vandenbussche F, Hu Y, Philips W, Van Der Straeten D (2012) Rosette tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiol 160:1149–1159
Deery DM, Rebetzke GJ, Jimenez-Berni JA, James RA, Condon AG, Bovill WD, Hutchinson P, Scarrow J, Davy R, Furbank RT (2016) Methodology for high-throughput field phenoty** of canopy temperature using airborne thermography. Front Plant Sci 7:1808–1808
Dungey HS, Dash JP, Pont D, Clinton PW, Watt MS, Telfer EJ (2018) Phenoty** whole forests will help to track genetic performance. Trends Plant Sci 23:854–864
Dutta P, Bandopadhyay P, Bera AK (2016) Identification of leaf based physiological markers for drought susceptibility during early seedling development of mungbean. Am J Plant Sci 7:1921
Fahlgren N, Gehan MA, Baxter I (2015) Lights, camera, action: high-throughput plant phenoty** is ready for a close-up. Curr Opin Plant Biol 24:93–99
Fiorani F, Schurr U (2013) Future scenarios for plant phenoty**. Annu Rev Plant Biol 64:267–291
Flood PJ, Kruijer W, Schnabel SK, van der Schoor R, Jalink H, Snel JFH, Harbinson J, Aarts MGM (2016) Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods 12:1–14
Furbank RT, Tester M (2011) Phenomics–technologies to relieve the phenoty** bottleneck. Trends Plant Sci 16:635–644
Furbank RT, Jimenez-Berni JA, George-Jaeggli B, Potgieter AB, Deery DM (2019) Field crop phenomics: enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol 223:1714–1727
Gage JL, White MR, Edwards JW, Kaeppler S, de Leon N (2018) Selection signatures underlying dramatic male inflorescence transformation during modern hybrid maize breeding. Genetics 210:1125–1138
Granier C, Aguirrezabal L, Chenu K, Cookson SJ, Dauzat M, Hamard P, Thioux JJ, Rolland G, Bouchier-Combaud S, Lebaudy A (2006) PHENOPSIS, an automated platform for reproducible phenoty** of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol 169:623–635
Guo D, Juan J, Chang L, Zhang J, Huang D (2017) Discrimination of plant root zone water status in greenhouse production based on phenoty** and machine learning techniques. Sci Rep 7:1–12
Guo Z, Yang W, Chang Y, Ma X, Tu H, **ong F, Jiang N, Feng H, Huang C, Yang P (2018) Genome-wide association studies of image traits reveal genetic architecture of drought resistance in rice. Mol Plant 11:789–805
Hatfield JL, Prueger JH (2015) Temperature extremes: effect on plant growth and development. Weather Clim Extremes 10:4–10
Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal-Bertioli SCM, Tester M, Gao C, Godwin ID, Hayes BJ, Wulff BBH (2019) Breeding crops to feed 10 billion. Nat Biotechnol 37:744–754
Houle D, Govindaraju DR, Omholt S (2010) Phenomics: the next challenge. Nat Rev Genet 11:855–866
Howarth CJ, Gay AP, Draper J, Powell W (2011) Development of high throughput plant phenoty** facilities at Aberystwyth. In: Proceedings of the phenomics workshop, San Diego
Humplík JF, Lazár D, Fürst T, Husičková A, Hýbl M, Spíchal L (2015) Automated integrative high-throughput phenoty** of plant shoots: a case study of the cold-tolerance of pea (Pisum sativum L.). Plant Methods 11:1–11
Hairmansis, A, Berger B, Tester M, Roy SJ (2014) Image-based phenoty** for non-destructive screening of different salinity tolerance traits in rice. Rice 7(1):1-10
Jansen M, Gilmer F, Biskup B, Nagel KA, Rascher U, Fischbach A, Briem S, Dreissen G, Tittmann S, Braun S (2009) Simultaneous phenoty** of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Functional Plant Biology 36(11):902–914
Jensen T, Apan A, Young F, Zeller L (2007) Detecting the attributes of a wheat crop using digital imagery acquired from a low-altitude platform. Comput Electron Agric 59:66–77
Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito M-I, Lin X (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Natu 408(6814):796–815
Klukas C, Chen D, Pape J-M (2014) Integrated analysis platform: an open-source information system for high-throughput plant phenoty**. Plant Physiol 165:506–518
Kruse OMO, Prats-Montalbán JM, Indahl UG, Kvaal K, Ferrer A, Futsaether CM (2014) Pixel classification methods for identifying and quantifying leaf surface injury from digital images. Comput Electron Agric 108:155–165
Langstroff A, Heuermann MC, Stahl A, Junker A (2021) Opportunities and limits of controlled-environment plant phenoty** for climate response traits. Theor Appl Genet 135(1):1–16
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444
Leiboff S, Li X, Hu HC, Todt N, Yang J, Li X, Yu X, Muehlbauer GJ, Timmermans MC, Yu J, Schnable PS, Scanlon MJ (2015) Genetic control of morphometric diversity in the maize shoot apical meristem. Nat Commun 6:8974
Leport L, Musse M, Cambert M, De Franscesci L, Le Caherec F, Burel A, Mariette F, Bouchereau A (2011) Canola leaf senescence phenoty** and identification of subcellular changes using NMR tool. In: 2nd international plant phenoty** symposium toward plant phenoty** science: challenges and perspectives, Forschungszentrum Jülich, Germany, pp 13
Li L, Zhang Q, Huang D (2014) A review of imaging techniques for plant phenoty**. Sensors 14:20078–20111
Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association map** of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc Natl Acad Sci 107:21199–21204
Li Y, **ao J, Chen L, Huang X, Cheng Z, Han B, Zhang Q, Wu C (2018) Rice functional genomics research: past decade and future. Mol Plant 11:359–380
Lymperopoulos P, Msanne J, Rabara R (2018) Phytochrome and phytohormones: working in Tandem for plant growth and development. Front Plant Sci 9:1037–1037
Mahlein AK, Kuska MT, Behmann J, Polder G, Walter A (2018) Hyperspectral sensors and imaging technologies in phytopathology: state of the art. Annu Rev Phytopathol 56:535–558
Minamikawa MF, Nonaka K, Kaminuma E, Kajiya-Kanegae H, Onogi A, Goto S, Yoshioka T, Imai A, Hamada H, Hayashi T (2017) Genome-wide association study and genomic prediction in citrus: potential of genomics-assisted breeding for fruit quality traits. Sci Rep 7:1–13
Montes JM, Utz HF, Schipprack W, Kusterer B, Muminovic J, Paul C, Melchinger AE (2006) Near-infrared spectroscopy on combine harvesters to measure maize grain dry matter content and quality parameters. Plant Breed 125:591–595
Montes JM, Melchinger AE, Reif JC (2007) Novel throughput phenoty** platforms in plant genetic studies. Trends Plant Sci 12:433–436
Mukherjee S, Mishra A, Trenberth KE (2018) Climate change and drought: a perspective on drought indices. Curr Clim Change Rep 4:145–163
Mula MG, Patil SB, Aden J, Rathore A, Anil Kumar V, Kumar RV (2016) Screening of pigeonpea genotypes for drought stress at early vegetative phase in Alfisol and Vertisol. Green Farming 7:507–511
Muraya MM, Chu J, Zhao Y, Junker A, Klukas C, Reif JC, Altmann T (2017) Genetic variation of growth dynamics in maize (Zea mays L.) revealed through automated non-invasive phenoty**. Plant J 89:366–380
Nabwire S, Suh H-K, Kim MS, Baek I, Cho B-K (2021) Application of artificial intelligence in phenomics. Sensors 21:4363
Nagel KA, Putz A, Gilmer F, Heinz K, Fischbach A, Pfeifer J, Faget M, Blossfeld S, Ernst M, Dimaki C (2012) GROWSCREEN-Rhizo is a novel phenoty** robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol 39:891–904
Neumann K, Klukas C, Friedel S, Rischbeck P, Chen D, Entzian A, Stein N, Graner A, Kilian B (2015) Dissecting spatiotemporal biomass accumulation in barley under different water regimes using high-throughput image analysis. Plant Cell Environ 38:1980–1996
Neumann K, Zhao Y, Chu J, Keilwagen J, Reif JC, Kilian B, Graner A (2017) Genetic architecture and temporal patterns of biomass accumulation in spring barley revealed by image analysis. BMC Plant Biol 17:1–12
Obidiegwu JE, Bryan GJ, Jones HG, Prashar A (2015) Co** with drought: stress and adaptive responses in potato and perspectives for improvement. Front Plant Sci 6:542
Oerke E-C, Steiner U, Dehne H-W, Lindenthal M (2006) Thermal imaging of cucumber leaves affected by downy mildew and environmental conditions. J Exp Bot 57:2121–2132
Parent B, Shahinnia F, Maphosa L, Berger B, Rabie H, Chalmers K, Kovalchuk A, Langridge P, Fleury D (2015) Combining field performance with controlled environment plant imaging to identify the genetic control of growth and transpiration underlying yield response to water-deficit stress in wheat. J Exp Bot 66:5481–5492
Pasala R, Pandey BB (2020) Plant phenomics: high-throughput technology for accelerating genomics. J Biosci 45:1–6
Patanè C, Scordia D, Testa G, Cosentino SL (2016) Physiological screening for drought tolerance in Mediterranean long-storage tomato. Plant Sci 249:25–34
Pereyra-Irujo GA, Gasco ED, Peirone LS, Aguirrezábal LAN (2012) GlyPh: a low-cost platform for phenoty** plant growth and water use. Funct Plant Biol 39:905–913
Poorter H, Fiorani F, Pieruschka R, Wojciechowski T, van der Putten WH, Kleyer M, Schurr U, Postma J (2016) Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol 212:838–855
Pound MP, Atkinson JA, Townsend AJ, Wilson MH, Griffiths M, Jackson AS, Bulat A, Tzimiropoulos G, Wells DM, Murchie EH (2017) Deep machine learning provides state-of-the-art performance in image-based plant phenoty**. Gigascience 6:gix083
Pratap A, Gupta S, Nair RM, Gupta SK, Schafleitner R, Basu PS, Singh CM, Prajapati U, Gupta AK, Nayyar H, Mishra AK, Baek K-H (2019) Using plant phenomics to exploit the gains of genomics. Agronomy 9
Rasheed A, **a X, Ogbonnaya F, Mahmood T, Zhang Z, Mujeeb-Kazi A, He Z (2014) Genome-wide association for grain morphology in synthetic hexaploid wheats using digital imaging analysis. BMC Plant Biol 14:1–21
Rebolledo MC, Peña AL, Duitama J, Cruz DF, Dingkuhn M, Grenier C, Tohme J (2016) Combining image analysis, genome wide association studies and different field trials to reveal stable genetic regions related to panicle architecture and the number of spikelets per panicle in rice. Front Plant Sci 7:1384
Reuzeau C, Pen J, Frankard V, de Wolf J, Peerbolte R, Broekaert WF, van Wim C (2010) TraitMill: a discovery engine for identifying yield-enhancement genes in cereals. Plant Gene Trait
Roscher R, Bohn B, Duarte MF, Garcke J (2020) Explainable machine learning for scientific insights and discoveries. Ieee Access 8:42200–42216
Rosenzweig C, Elliott J, Deryng D, Ruane AC, Müller C, Arneth A, Boote KJ, Folberth C, Glotter M, Khabarov N (2014) Assessing agricultural risks of climate change in the 21st century in a global gridded crop model intercomparison. Proc Natl Acad Sci 111:3268–3273
Ruiz-Garcia L, Lunadei L, Barreiro P, Robla I (2009) A review of wireless sensor technologies and applications in agriculture and food industry: state of the art and current trends. Sensors 9:4728–4750
Sabanci K, Kayabasi A, Toktas A (2017) Computer vision-based method for classification of wheat grains using artificial neural network. J Sci Food Agric 97:2588–2593
Sadok W, Naudin P, Boussuge B, Muller B, Welcker C, Tardieu F (2007) Leaf growth rate per unit thermal time follows QTL-dependent daily patterns in hundreds of maize lines under naturally fluctuating conditions. Plant Cell Environ 30:135–146
Shi C, Zhao L, Zhang X, Lv G, Pan Y, Chen F (2019) Gene regulatory network and abundant genetic variation play critical roles in heading stage of polyploidy wheat. BMC Plant Biol 19:1–16
Shi L, Shi T, Broadley MR, White PJ, Long Y, Meng J, Xu F, Hammond JP (2013) High-throughput root phenoty** screens identify genetic loci associated with root architectural traits in Brassica napus under contrasting phosphate availabilities. Ann Bot 112:381–389
Singh AK, Ganapathysubramanian B, Sarkar S, Singh A (2018) Deep learning for plant stress phenoty**: trends and future perspectives. Trends Plant Sci 23:883–898
Sirault X, Fripp J, Paproki A, Kuffner P, Nguyen C, Li R, Daily H, Guo J, Furbank R (2013) PlantScan: a three-dimensional phenoty** platform for capturing the structural dynamic of plant development and growth. In: Proceedings of the 7th international conference on functionalstructural plant models, Saariselk, Finland, pp 45–48
Sterck L, Rombauts S, Vandepoele K, Rouzé P, Van de Peer Y (2007) How many genes are there in plants (… and why are they there)? Curr Opin Plant Biol 10:199–203
Taylor JF (2014) Implementation and accuracy of genomic selection. Aquaculture 420:S8–S14
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
Tisné S, Serrand Y, Bach L, Gilbault E, Ben Ameur R, Balasse H, Voisin R, Bouchez D, Durand-Tardif M, Guerche P, Chareyron G, Da Rugna J, Camilleri C, Loudet O (2013) Phenoscope: an automated large-scale phenoty** platform offering high spatial homogeneity. Plant J 74:534–544
Vadez V, Kholová J, Hummel G, Zhokhavets U, Gupta SK, Hash CT (2015) LeasyScan: a novel concept combining 3D imaging and lysimetry for high-throughput phenoty** of traits controlling plant water budget. J Exp Bot 66:5581–5593
Van Raan AFJ (2004) Measuring science, Handbook of quantitative science and technology research. Springer, pp 19–50
Virlet N, Sabermanesh K, Sadeghi-Tehran P, Hawkesford MJ (2016) Field Scanalyzer: an automated robotic field phenoty** platform for detailed crop monitoring. Functional Plant Bio 44(1):143–153.
Walter T, Shattuck DW, Baldock R, Bastin ME, Carpenter AE, Duce S, Ellenberg J, Fraser A, Hamilton N, Pieper S (2010) Visualization of image data from cells to organisms. Nat Methods 7:S26–S41
Walter A, Scharr H, Gilmer F, Zierer R, Nagel KA, Ernst M, Wiese A, Virnich O, Christ MM, Uhlig B (2007) Dynamics of seedling growth acclimation towards altered light conditions can be quantified via GROWSCREEN: a setup and procedure designed for rapid optical phenoty** of different plant species. New Phytologist 174(2):447–455
Wang P, Zhou G, Yu H, Yu S (2011) Fine map** a major QTL for flag leaf size and yield-related traits in rice. Theor Appl Genet 123:1319–1330
Wang Q, **e W, **ng H, Yan J, Meng X, Li X, Fu X, Xu J, Lian X, Yu S (2015) Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol Plant 8:946–957
Wang X, Zhang R, Song W, Han L, Liu X, Sun X, Luo M, Chen K, Zhang Y, Yang H (2019) Dynamic plant height QTL revealed in maize through remote sensing phenoty** using a high-throughput unmanned aerial vehicle (UAV). Sci Rep 9:1–10
Waring RH, Cleary BD (1967) Plant moisture stress: evaluation by pressure bomb. Science 155:1248–1254
Weight C, Parnham D, Waites R (2008) TECHNICAL ADVANCE: LeafAnalyser: a computational method for rapid and large-scale analyses of leaf shape variation. Plant J 53:578–586
Wetterich CB, Kumar R, Sankaran S, Junior JB, Ehsani R, Marcassa LG A comparative study on application of computer vision and fluorescence imaging spectroscopy for detection of citrus huanglongbing disease in USA and Brazil. Opt Soc Am: JW3A–JW26A
Wu D, Guo Z, Ye J, Feng H, Liu J, Chen G, Zheng J, Yan D, Yang X, ** and genome-wide association study to dissect the genetic architecture of tiller growth in rice. J Exp Bot 70:545–561
Wu X et al (2021) Using high-throughput multiple optical phenoty** to decipher the genetic architecture of maize drought tolerance. Genome Biol 22:185
Wu H, Jaeger M, Wang M, Li B, Zhang BG (2011) Three-dimensional distribution of vessels, passage cells and lateral roots along the root axis of winter wheat (Triticum aestivum). Ann of Bot 107(5):843–853
**ao Y, Liu H, Wu L, Warburton M, Yan J (2017) Genome-wide association studies in maize: praise and stargaze. Mol Plant 10:359–374
Xue B, Sartori P, Leibler S (2019) Environment-to-phenotype map** and adaptation strategies in varying environments. Proc Natl Acad Sci 116:13847–13855
Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, Fang W, Feng H, ** and genome-wide association studies to reveal natural genetic variation in rice. Nat Commun 5:1–9
Yang W, Guo Z, Huang C, Wang K, Jiang N, Feng H, Chen G, Liu Q, **ong L (2015) Genome-wide association study of rice (Oryza sativa L.) leaf traits with a high-throughput leaf scorer. J Exp Bot 66:5605–5615
Yang W, Feng H, Zhang X, Zhang J, Doonan JH, Batchelor WD, **: past decades, current challenges, and future perspectives. Mol Plant 13:187–214
Yang Y, Chai R, He Y (2012) Early detection of rice blast (Pyricularia) at seedling stage in Nipponbare rice variety using near-infrared hyper-spectral image. Afr J Biotechnol 11:6809–6817
Yao W, Li G, Yu Y, Ouyang Y (2018) funRiceGenes dataset for comprehensive understanding and application of rice functional genes. Gigascience 7:1–9
Zakaluk R, Ranjan R (2008) Predicting the leaf water potential of potato plants using RGB reflectance. Can Biosyst Eng 50
Zarco-Tejada PJ, Berni JAJ, Suárez L, Sepulcre-Cantó G, Morales F, Miller JR (2009) Imaging chlorophyll fluorescence with an airborne narrow-band multispectral camera for vegetation stress detection. Remote Sens Environ 113:1262–1275
Zhang C, Pumphrey M, Zhou J, Gao H, Zhang Q, Sankaran S (2017) Development of automated highthroughput phenoty** system for controlled environment studies, Paper No. 1700581, 2017 American Society of Agricultural and Biological Engineers (ASABE) Annual International Meeting (AIM), Spokane, WA
Zhang Q, Li J, Xue Y, Han B, Deng XW (2008) Rice 2020: a call for an international coordinated effort in rice functional genomics. Mol Plant 1:715–719
Zhao C, Liu B, Piao S, Wang X, Lobell DB, Huang Y, Huang M, Yao Y, Bassu S, Ciais P (2017) Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci 114:9326–9331
Zhao C, Zhang Y, Du J, Guo X, Wen W, Gu S, Wang J, Fan J (2019) Crop phenomics: current status and perspectives. Front Plant Sci:10
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Waseem, M., Shaheen, I., Aslam, M.M. (2022). Advances in Integrated High-Throughput and Phenomics Application in Plants and Agriculture. In: Prakash, C.S., Fiaz, S., Fahad, S. (eds) Principles and Practices of OMICS and Genome Editing for Crop Improvement. Springer, Cham. https://doi.org/10.1007/978-3-030-96925-7_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-96925-7_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-96924-0
Online ISBN: 978-3-030-96925-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)