Abstract
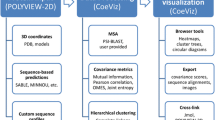
CEvADA is a database of amino acid coevolution networks aimed to detect specificity determinant and function related sites in protein families. The database was also designed to provide an easy access to protein coevolutionary constraints that can be incorporated in machine learning classification models, just as sequence annotation and structure prediction methods. The data can be accessed for the whole protein family and specific protein sequences. We also provide sequence search and a REST API for programmatic access in the database. The current version of the database contains data related to 6.301 conserved domains and 45 million protein sequences. CeVADA is free and can be accessed at http://bioinfo.icb.ufmg.br/cevada.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Almende, B., Benoit, T., Titouan, R.: Package ‘visnetwork’. Netw. Visu. ‘vis. js’ Lib. Vers. 2(9) (2019). https://mran.microsoft.com/snapshot/2016-10-12/web/packages/visNetwork/visNetwork.pdf
Bachega, J.F.R., et al.: Systematic structural studies of iron superoxide dismutases from human parasites and a statistical coupling analysis of metal binding specificity. Proteins: Struct. Funct. Bioinf. 77(1), 26–37 (2009). https://doi.org/10.1002/prot.22412
Barwinska-Sendra, A., et al.: An evolutionary path to altered cofactor specificity in a metalloenzyme. Nat. Commun. 11(1), 1–13 (2020). https://doi.org/10.1038/s41467-020-16478-0
Bostock, M., Ogievetsky, V., Heer, J.: D\(^3\) data-driven documents. IEEE Trans. Vis. Comput. Graph. 17(12), 2301–2309 (2011). https://doi.org/10.1109/TVCG.2011.185
Chakraborty, A., Chakrabarti, S.: A survey on prediction of specificity-determining sites in proteins. Briefings Bioinf. 16(1), 71–88 (2015). https://doi.org/10.1093/bib/bbt092
Choi, Y., Sims, G.E., Murphy, S., Miller, J.R., Chan, A.P.: Predicting the functional effect of amino acid substitutions and indels. PloS one 7(10), e46688 (2012). https://doi.org/10.1371/journal.pone.0046688
Coitinho, J.B., et al.: Structural and immunological characterization of a new nucleotidyltransferase-like antigen from Paracoccidioides brasiliensis. Mol. Immunol. 112, 151–162 (2019). https://doi.org/10.1016/j.molimm.2019.04.028
El-Gebali, S., et al.: The Pfam protein families database in 2019. Nucleic Acids Res. 47(D1), D427–D432 (2019). https://doi.org/10.1093/nar/gky995
da Fonseca, N.J., Afonso, M.Q.L., de Oliveira, L.C., Bleicher, L.: A new method bridging graph theory and residue co-evolutionary networks for specificity determinant positions detection. Bioinformatics 35(9), 1478–1485 (2019). https://doi.org/10.1093/bioinformatics/bty846
Fonseca, N., Afonso, M., Carrijo, L., Bleicher, L.: Conan: a web application to detect specificity determinants and functional sites by amino acids co-variation network analysis. Bioinformatics (2020). https://doi.org/10.1093/bioinformatics/btaa713
da Fonseca, N.J., Afonso, M.Q.L., Pedersolli, N.G., de Oliveira, L.C., Andrade, D.S., Bleicher, L.: Sequence, structure and function relationships in flaviviruses as assessed by evolutive aspects of its conserved non-structural protein domains. Biochem. Biophys. Res. Commun. 492(4), 565–571 (2017). https://doi.org/10.1016/j.bbrc.2017.01.041
Jaccard, P.: The distribution of the flora in the alpine zone. 1. New Phytol. 11(2), 37–50 (1912).https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Lima Afonso, M., de Lima, L., Bleicher, L.: Residue correlation networks in nuclear receptors reflect functional specialization and the formation of the nematode-specific P-box. BMC Genomics 14(Suppl 6), S1 (2013)
Lockless, S.W., Ranganathan, R.: Evolutionarily conserved pathways of energetic connectivity in protein families. Science 286(5438), 295–299 (1999). https://doi.org/10.1126/science.286.5438.295
Oliveira, A., Bleicher, L., Schrago, C.G., Junior, F.P.S.: Conservation analysis and decomposition of residue correlation networks in the phospholipase a2 superfamily (pla2s): Insights into the structure-function relationships of snake venom toxins. Toxicon 146, 50–60 (2018). https://doi.org/10.1016/j.toxicon.2018.03.013
Querino Lima Afonso, M., da Fonseca, N.J., de Oliveira, L.C., Lobo, F.P., Bleicher, L.: Coevolved positions represent key functional properties in the trypsin-like serine proteases protein family. J. Chem. Inf. Model. 60(2), 1060–1068 (2020). https://doi.org/10.1021/acs.jcim.9b00903
Rauer, C., Sen, N., Waman, V.P., Abbasian, M., Orengo, C.A.: Computational approaches to predict protein functional families and functional sites. Curr. Opin. Struct. Biol. 70, 108–122 (2021). https://doi.org/10.1016/j.sbi.2021.05.012
Rios-Anjos, R.M., de Lima Camandona, V., Bleicher, L., Ferreira-Junior, J.R.: Structural and functional map** of Rtg2p determinants involved in retrograde signaling and aging of Saccharomyces cerevisiae. PloS One 12(5) (2017). https://doi.org/10.1371/journal.pone.0177090
Taylor, W.R.: The classification of amino acid conservation. J. Theor. Biol. 119(2), 205–218 (1986). https://doi.org/10.1016/s0022-5193(86)80075-3
Tumminello, M., Miccichè, S., Lillo, F., Piilo, J., Mantegna, R.N.: Statistically validated networks in bipartite complex systems. PLoS ONE 6(3) (2011). https://doi.org/10.1371/journal.pone.0017994
Watkins, X., Garcia, L.J., Pundir, S., Martin, M.J., Consortium, U.: Protvista: visualization of protein sequence annotations. Bioinformatics 33(13), 2040–2041 (2017). https://doi.org/10.1093/bioinformatics/btx120
Yachdav, G., et al.: Msaviewer: interactive javascript visualization of multiple sequence alignments. Bioinformatics 32(22), 3501–3503 (2016). https://doi.org/10.1093/bioinformatics/btw474
Zuckerkandl, E., Pauling, L.: Evolutionary divergence and convergence in proteins. In: Evolving Genes and Proteins, pp. 97–166. Elsevier (1965). https://doi.org/10.1016/B978-1-4832-2734-4.50017-6
Acknowledgements
Authors thank CEPAD-ICB, UFMG, for hosting CeVADA in the HPC cluster Sagarana.
Funding
This work has been supported by CNPq (grant 457851/2014-7) and CAPES (grant 051/2013).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
da Fonseca Júnior, N.J., Afonso, M.Q.L., Bleicher, L. (2021). CEvADA: Co-Evolution Analysis Data Archive. In: Stadler, P.F., Walter, M.E.M.T., Hernandez-Rosales, M., Brigido, M.M. (eds) Advances in Bioinformatics and Computational Biology. BSB 2021. Lecture Notes in Computer Science(), vol 13063. Springer, Cham. https://doi.org/10.1007/978-3-030-91814-9_11
Download citation
DOI: https://doi.org/10.1007/978-3-030-91814-9_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91813-2
Online ISBN: 978-3-030-91814-9
eBook Packages: Computer ScienceComputer Science (R0)