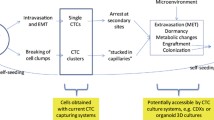
Abstract
Metastasis is the major cause of breast cancer death worldwide. In metastatic breast cancer, circulating tumor cells (CTCs) can be captured from patient blood samples sequentially over time and thereby serve as surrogates to assess the biology of surviving cancer cells that may still persist in solitary or multiple metastatic sites following treatment. CTCs may thus function as potential real-time decision-making guides for selecting appropriate therapies during the course of disease or for the development and testing of new treatments. The heterogeneous nature of CTCs warrants the use of single cell platforms to better inform our understanding of these cancer cells. Current techniques for single cell analyses and techniques for investigating interactions between cancer and immune cells are discussed. In addition, methodologies for growing patient-derived CTCs in vitro or propagating them in vivo to facilitate CTC drug testing are reviewed. We advocate the use of CTCs in appropriate microenvironments to appraise the effectiveness of cancer chemotherapies, immunotherapies, and for the development of new cancer treatments, fundamental to personalizing and improving the clinical management of metastatic breast cancer.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. https://doi.org/10.3322/caac.21551.
Malvezzi M, Carioli G, Bertuccio P, Boffetta P, Levi F, La Vecchia C, et al. European cancer mortality predictions for the year 2019 with focus on breast cancer. Ann Oncol. 2019;30(5):781–7. https://doi.org/10.1093/annonc/mdz051.
Carioli G, Malvezzi M, Rodriguez T, Bertuccio P, Negri E, La Vecchia C. Trends and predictions to 2020 in breast cancer mortality: Americas and Australasia. Breast (Edinburgh, Scotland). 2018;37:163–9. https://doi.org/10.1016/j.breast.2017.12.004.
Mariotto AB, Etzioni R, Hurlbert M, Penberthy L, Mayer M. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomark Prev. 2017;26(6):809–15. https://doi.org/10.1158/1055-9965.EPI-16-0889.
Sledge GW Jr. Curing metastatic breast cancer. J Oncol Prac. 2016;12(1):6–10. https://doi.org/10.1200/JOP.2015.008953.
Hattori M, Iwata H. Advances in treatment and care in metastatic breast cancer (MBC): are there MBC patients who are curable? Chin Clin Oncol. 2018;7(3):23. https://doi.org/10.21037/cco.2018.05.01.
Guth U, Elfgen C, Montagna G, Schmid SM. Long-term survival and cure in distant metastatic breast cancer. Oncology. 2019:1–12. https://doi.org/10.1159/000500298.
den Brok WD, Speers CH, Gondara L, Baxter E, Tyldesley SK, Lohrisch CA. Survival with metastatic breast cancer based on initial presentation, de novo versus relapsed. Breast Cancer Res Treat. 2017;161(3):549–56. https://doi.org/10.1007/s10549-016-4080-9.
Klar N, Rosenzweig M, Diergaarde B, Brufsky A. Features associated with long-term survival in patients with metastatic breast cancer. Clin Breast Cancer. 2019;19(4):304–10. https://doi.org/10.1016/j.clbc.2019.01.014.
Tong CWS, Wu M, Cho WCS, To KKW. Recent advances in the treatment of breast cancer. Front Oncol. 2018;8:227. https://doi.org/10.3389/fonc.2018.00227.
Gobbini E, Ezzalfani M, Dieras V, Bachelot T, Brain E, Debled M, et al. Time trends of overall survival among metastatic breast cancer patients in the real-life ESME cohort. Eur J Cancer. 2018;96:17–24. https://doi.org/10.1016/j.ejca.2018.03.015.
Perol D, Robain M, Arveux P, Mathoulin-Pelissier S, Chamorey E, Asselain B, et al. The ongoing French metastatic breast cancer (MBC) cohort: the example-based methodology of the Epidemiological Strategy and Medical Economics (ESME). BMJ Open. 2019;9(2):e023568. https://doi.org/10.1136/bmjopen-2018-023568.
Flowers M, Birkey Reffey S, Mertz SA, Hurlbert M, for the Metastatic Breast Cancer Alliance. Obstacles, opportunities and priorities for advancing metastatic breast cancer research. Cancer Res. 2017;77(13):3386–90. https://doi.org/10.1158/0008-5472.CAN-17-0232.
Parry M. Introducing the Metastatic Breast Cancer Project: a novel patient-partnered initiative to accelerate understanding of MBC. ESMO Open. 2018;3(7):e000452. https://doi.org/10.1136/esmoopen-2018-000452.
Friedman R. Drug resistance in cancer: molecular evolution and compensatory proliferation. Oncotarget. 2016;7(11):11746–55. https://doi.org/10.18632/oncotarget.7459.
McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168(4):613–28. https://doi.org/10.1016/j.cell.2017.01.018.
Dupont Jensen J, Laenkholm A-V, Knoop A, Ewertz M, Bandaru R, Liu W, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17(4):667–77. https://doi.org/10.1158/1078-0432.CCR-10-1133.
Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. https://doi.org/10.1056/NEJMoa1113205.
de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346(6206):251–6. https://doi.org/10.1126/science.1253462.
Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9. https://doi.org/10.1126/science.1256930.
Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the evolution of non-small-cell lung cancer. N Engl J Med. 2017;376(22):2109–21. https://doi.org/10.1056/NEJMoa1616288.
Zou S-M, Li W-H, Wang W-M, Li W-B, Shi S-S, Ying J-M, et al. The gene mutational discrepancies between primary and paired metastatic colorectal carcinoma detected by next-generation sequencing. J Cancer Res Clin Oncol. 2018;144(11):2149–59. https://doi.org/10.1007/s00432-018-2742-1.
Wu JM, Fackler MJ, Halushka MK, Molavi DW, Taylor ME, Teo WW, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14(7):1938–46. https://doi.org/10.1158/1078-0432.CCR-07-4082.
Kroigard AB, Larsen MJ, Thomassen M, Kruse TA. Molecular concordance between primary breast cancer and matched metastases. Breast J. 2016;22(4):420–30. https://doi.org/10.1111/tbj.12596.
Cummings MC, Simpson PT, Reid LE, Jayanthan J, Skerman J, Song S, et al. Metastatic progression of breast cancer: insights from 50 years of autopsies. J Pathol. 2014;232(1):23–31. https://doi.org/10.1002/path.4288.
Avigdor BE, Cimino-Mathews A, DeMarzo AM, Hicks JL, Shin J, Sukumar S et al. Mutational profiles of breast cancer metastases from a rapid autopsy series reveal multiple evolutionary trajectories. JCI Insight 2017;2(24). pii: 96896. https://doi.org/10.1172/jci.insight.96896.
Navin N, Krasnitz A, Rodgers L, Cook K, Meth J, Kendall J, et al. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20(1):68–80. https://doi.org/10.1101/gr.099622.109.
Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736-49.e18. https://doi.org/10.1016/j.cell.2017.04.016.
Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984-97.e24. https://doi.org/10.1016/j.cell.2018.09.006.
Siebert JC, Walker EB. Monitoring cytokine profiles during immunotherapy. Immunotherapy. 2010;2(6):799–816. https://doi.org/10.2217/imt.10.76.
Kim C, Gao R, Sei E, Brandt R, Hartman J, Hatschek T, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879-93.e13. https://doi.org/10.1016/j.cell.2018.03.041.
Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016;537(7618):102–6. https://doi.org/10.1038/nature19328.
Miyamoto DT, Zheng Y, Wittner BS, Lee RJ, Zhu H, Broderick KT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349(6254):1351–6. https://doi.org/10.1126/science.aab0917.
Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–401. https://doi.org/10.1126/science.1254257.
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352(6282):189–96. https://doi.org/10.1126/science.aad0501.
Gong H, Wang X, Liu B, Boutet S, Holcomb I, Dakshinamoorthy G, et al. Single-cell protein-mRNA correlation analysis enabled by multiplexed dual-analyte co-detection. Sci Rep. 2017;7(1):2776. https://doi.org/10.1038/s41598-017-03057-5.
Chen Z, Lu Y, Zhang K, **ao Y, Lu J, Fan R. Multiplexed, sequential secretion analysis of the same single cells reveals distinct effector response dynamics dependent on the initial basal state. Adv Sci (Weinheim, Baden-Wurttemberg, Germany). 2019;6(9):1801361. https://doi.org/10.1002/advs.201801361.
Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2009;81(16):6813–22. https://doi.org/10.1021/ac901049w.
Giesen C, Wang HAO, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat Methods. 2014;11(4):417–22. https://doi.org/10.1038/nmeth.2869.
Levine JH, Simonds EF, Bendall SC, Davis KL, Amir EAD, Tadmor MD, et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell. 2015;162(1):184–97. https://doi.org/10.1016/j.cell.2015.05.047.
Good Z, Sarno J, Jager A, Samusik N, Aghaeepour N, Simonds EF, et al. Single-cell developmental classification of B cell precursor acute lymphoblastic leukemia at diagnosis reveals predictors of relapse. Nat Med. 2018;24(4):474–83. https://doi.org/10.1038/nm.4505.
Gonzalez VD, Samusik N, Chen TJ, Savig ES, Aghaeepour N, Quigley DA, et al. Commonly occurring cell subsets in high-grade serous ovarian tumors identified by single-cell mass cytometry. Cell Rep. 2018;22(7):1875–88. https://doi.org/10.1016/j.celrep.2018.01.053.
Chew V, Lai L, Pan L, Lim CJ, Li J, Ong R, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci U S A. 2017;114(29):E5900–E5909. https://doi.org/10.1073/pnas.1706559114.
Lavin Y, Kobayashi S, Leader A, Amir EAD, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169(4):750-65.e17. https://doi.org/10.1016/j.cell.2017.04.014.
Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. 2019;176(1-2):98–112.e14. https://doi.org/10.1016/j.cell.2018.11.046.
Bian S, Hou Y, Zhou X, Li X, Yong J, Wang Y, et al. Single-cell multiomics sequencing and analyses of human colorectal cancer. Science. 2018;362(6418):1060–3. https://doi.org/10.1126/science.aao3791.
Rodriguez-Meira A, Buck G, Clark S-A, Povinelli BJ, Alcolea V, Louka E, et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell. 2019;73(6):1292-305.e8. https://doi.org/10.1016/j.molcel.2019.01.009.
Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One. 2012;7(5):e33788. https://doi.org/10.1371/journal.pone.0033788.
Tsuyuzaki K, Ishii M, Nikaido I. Uncovering hypergraphs of cell-cell interaction from single cell RNA-sequencing data. bioRxiv. 2019. https://doi.org/10.1101/566182.
Szczerba BM, Castro-Giner F, Vetter M, Krol I, Gkountela S, Landin J et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature. 2019;566(7745):553–7. https://doi.org/10.1038/s41586-019-0915-y.
Aceto N, Toner M, Maheswaran S, Haber DA. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer. 2015;1(1):44–52. https://doi.org/10.1016/j.trecan.2015.07.006.
Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107(43):18392–7. https://doi.org/10.1073/pnas.1012539107.
Lo Russo G, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G, et al. Antibody-Fc/FcR interaction on macrophages as a mechanism for hyperprogressive disease in non-small cell lung cancer subsequent to PD-1/PD-L1 blockade. Clin Cancer Res. 2019;25(3):989–99. https://doi.org/10.1158/1078-0432.CCR-18-1390.
Kumar MP, Du J, Lagoudas G, Jiao Y, Sawyer A, Drummond DC, et al. Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics. Cell Rep. 2018;25(6):1458-68.e4. https://doi.org/10.1016/j.celrep.2018.10.047.
Chen Y, Sun W, Kang L, Wang Y, Zhang M, Zhang H, et al. Microfluidic co-culture of liver tumor spheroids with stellate cells for the investigation of drug resistance and intercellular interactions. Analyst. 2019;144(14):4233–40. https://doi.org/10.1039/c9an00612e.
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865–8. https://doi.org/10.1038/nmeth.4380.
Peterson VM, Zhang KX, Kumar N, Wong J, Li L, Wilson DC, et al. Multiplexed quantification of proteins and transcripts in single cells. Nat Biotechnol. 2017;35(10):936–9. https://doi.org/10.1038/nbt.3973.
Todorovic V. Single-cell RNA-seq—now with protein. Nat Methods. 2017;14:1028–29. https://doi.org/10.1038/nmeth.4488.
Nishida-Aoki N, Gujral TS. Emerging approaches to study cell-cell interactions in tumor microenvironment. Oncotarget. 2019;10(7):785–97. https://doi.org/10.18632/oncotarget.26585.
Zhang L, Vertes A. Single-cell mass spectrometry approaches to explore cellular heterogeneity. Angew Chem Int Ed Engl. 2018;57(17):4466–77. https://doi.org/10.1002/anie.201709719.
Spitzer MH, Nolan GP. Mass cytometry: single cells, many features. Cell. 2016;165(4):780–91. https://doi.org/10.1016/j.cell.2016.04.019.
Rinkenbaugh AL, Sinha VC, Zhang X, Piwnica-Worms H. Abstract 2181: Functionalizing intratumoral signaling heterogeneity in triple negative breast cancer. Cancer Res. 2018;78(13 Supplement):2181. https://doi.org/10.1158/1538-7445.AM2018-2181.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10(12):1213–8. https://doi.org/10.1038/nmeth.2688.
Chen X, Shen Y, Draper W, Buenrostro JD, Litzenburger U, Cho SW, et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods. 2016;13(12):1013–20. https://doi.org/10.1038/nmeth.4031.
Huang TH, Velho T, Lois C. Monitoring cell-cell contacts in vivo in transgenic animals. Development. 2016;143(21):4073–84. https://doi.org/10.1242/dev.142406.
Gabrilovich DI. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5(1):3–8. https://doi.org/10.1158/2326-6066.CIR-16-0297.
Bruno A, Mortara L, Baci D, Noonan DM, Albini A. Myeloid derived suppressor cells interactions with natural killer cells and pro-angiogenic activities: roles in tumor progression. Front Immunol. 2019;10:771. https://doi.org/10.3389/fimmu.2019.00771.
Sprouse ML, Welte T, Boral D, Liu HN, Yin W, Vishnoi M et al. PMN-MDSCs enhance CTC metastatic properties through reciprocal interactions via ROS/Notch/Nodal signaling. Int J Mol Sci. 2019;20(8). pii: E1916. https://doi.org/10.3390/ijms20081916.
Kuznetsov HS, Marsh T, Markens BA, Castano Z, Greene-Colozzi A, Hay SA, et al. Identification of luminal breast cancers that establish a tumor-supportive macroenvironment defined by proangiogenic platelets and bone marrow-derived cells. Cancer Discov. 2012;2(12):1150–65. https://doi.org/10.1158/2159-8290.CD-12-0216.
Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell. 2015;28(5):666–76. https://doi.org/10.1016/j.ccell.2015.09.018.
Best MG, Wesseling P, Wurdinger T. Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res. 2018;78(13):3407–12. https://doi.org/10.1158/0008-5472.CAN-18-0887.
Gillet J-P, Varma S, Gottesman MM. The clinical relevance of cancer cell lines. J Natl Cancer Inst. 2013;105(7):452–8. https://doi.org/10.1093/jnci/djt007.
Wang R, Chu GCY, Mrdenovic S, Annamalai AA, Hendifar AE, Nissen NN, et al. Cultured circulating tumor cells and their derived xenografts for personalized oncology. Asian J Urol. 2016;3(4):240–53. https://doi.org/10.1016/j.ajur.2016.08.005.
den Toonder J. Circulating tumor cells: the Grand Challenge. Lab Chip. 2011;11(3):375–7. https://doi.org/10.1039/c0lc90100h.
Maheswaran S, Haber DA. Ex vivo culture of CTCs: an emerging resource to guide cancer therapy. Cancer Res. 2015;75(12):2411–5. https://doi.org/10.1158/0008-5472.CAN-15-0145.
Drost J, Clevers H. Organoids in cancer research. Nat Rev Cancer. 2018;18(7):407–18. https://doi.org/10.1038/s41568-018-0007-6.
Tuveson D, Clevers H. Cancer modeling meets human organoid technology. Science. 2019;364(6444):952–5. https://doi.org/10.1126/science.aaw6985.
Sachs N, de Ligt J, Kopper O, Gogola E, Bounova G, Weeber F, et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell. 2018;172(1–2):373-86.e10. https://doi.org/10.1016/j.cell.2017.11.010.
Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, et al. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell. 2018;174(6):1586-98.e12. https://doi.org/10.1016/j.cell.2018.07.009.
Gao D, Vela I, Sboner A, Iaquinta PJ, Karthaus WR, Gopalan A, et al. Organoid cultures derived from patients with advanced prostate cancer. Cell. 2014;159(1):176–87. https://doi.org/10.1016/j.cell.2014.08.016.
Yu M, Bardia A, Aceto N, Bersani F, Madden MW, Donaldson MC, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216–20. https://doi.org/10.1126/science.1253533.
Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20(8):897–903. https://doi.org/10.1038/nm.3600.
Paris PL, Kobayashi Y, Zhao Q, Zeng W, Sridharan S, Fan T, et al. Functional phenoty** and genoty** of circulating tumor cells from patients with castration resistant prostate cancer. Cancer Lett. 2009;277(2):164–73. https://doi.org/10.1016/j.canlet.2008.12.007.
Bobek V, Gurlich R, Eliasova P, Kolostova K. Circulating tumor cells in pancreatic cancer patients: enrichment and cultivation. World J Gastroenterol. 2014;20(45):17163–70. https://doi.org/10.3748/wjg.v20.i45.17163.
Bobek V, Kacprzak G, Rzechonek A, Kolostova K. Detection and cultivation of circulating tumor cells in malignant pleural mesothelioma. Anticancer Res. 2014;34(5):2565–9.
Bobek V, Matkowski R, Gurlich R, Grabowski K, Szelachowska J, Lischke R, et al. Cultivation of circulating tumor cells in esophageal cancer. Folia Histochem Cytobiol. 2014;52(3):171–7. https://doi.org/10.5603/FHC.2014.0020.
Cegan M, Kolostova K, Matkowski R, Broul M, Schraml J, Fiutowski M, et al. In vitro culturing of viable circulating tumor cells of urinary bladder cancer. Int J Clin Exp Pathol. 2014;7(10):7164–71.
Kolostova K, Broul M, Schraml J, Cegan M, Matkowski R, Fiutowski M, et al. Circulating tumor cells in localized prostate cancer: isolation, cultivation in vitro and relationship to T-stage and Gleason score. Anticancer Res. 2014;34(7):3641–6.
Kolostova K, Cegan M, Bobek V. Circulating tumour cells in patients with urothelial tumours: Enrichment and in vitro culture. Can Urol Assoc J. 2014;8(9–10):E715–20. https://doi.org/10.5489/cuaj.1978.
Zhang Z, Shiratsuchi H, Lin J, Chen G, Reddy RM, Azizi E, et al. Expansion of CTCs from early stage lung cancer patients using a microfluidic co-culture model. Oncotarget. 2014;5(23):12383–97. https://doi.org/10.18632/oncotarget.2592.
Kolostova K, Spicka J, Matkowski R, Bobek V. Isolation, primary culture, morphological and molecular characterization of circulating tumor cells in gynecological cancers. Am J Transl Res. 2015;7(7):1203–13.
Kolostova K, Matkowski R, Gurlich R, Grabowski K, Soter K, Lischke R, et al. Detection and cultivation of circulating tumor cells in gastric cancer. Cytotechnology. 2016;68(4):1095–102. https://doi.org/10.1007/s10616-015-9866-9.
Malara N, Trunzo V, Foresta U, Amodio N, De Vitis S, Roveda L, et al. Ex-vivo characterization of circulating colon cancer cells distinguished in stem and differentiated subset provides useful biomarker for personalized metastatic risk assessment. J Transl Med. 2016;14(1):133. https://doi.org/10.1186/s12967-016-0876-y.
Hwang E, Uh J-H, Lee HS, Lee CH, Lee SJ, Ahn SH, et al. Cancer gene panel analysis of cultured circulating tumor cells and primary tumor tissue from patients with breast cancer. Oncol Lett. 2017;13(6):4627–32. https://doi.org/10.3892/ol.2017.6077.
Bielcikova Z, Jakabova A, Pinkas M, Zemanova M, Kolostova K, Bobek V. Circulating tumor cells: what we know, what do we want to know about them and are they ready to be used in clinics? Am J Transl Res. 2017;9(6):2807–23.
Jakabova A, Bielcikova Z, Pospisilova E, Matkowski R, Szynglarewicz B, Staszek-Szewczyk U, et al. Molecular characterization and heterogeneity of circulating tumor cells in breast cancer. Breast Cancer Res Treat. 2017;166(3):695–700. https://doi.org/10.1007/s10549-017-4452-9.
Pizon M, Zimon D, Carl S, Pachmann U, Pachmann K, Camara O. Heterogeneity of circulating epithelial tumour cells from individual patients with respect to expression profiles and clonal growth (sphere formation) in breast cancer. Ecancermedicalscience. 2013;7:343. https://doi.org/10.3332/ecancer.2013.343.
Lu J, Fan T, Zhao Q, Zeng W, Zaslavsky E, Chen JJ, et al. Isolation of circulating epithelial and tumor progenitor cells with an invasive phenotype from breast cancer patients. Int J Cancer. 2010;126(3):669–83. https://doi.org/10.1002/ijc.24814.
Vishnoi M, Peddibhotla S, Yin W, Scamardo T, George GC, Hong DS, et al. The isolation and characterization of CTC subsets related to breast cancer dormancy. Sci Rep. 2015;5(1):17533. https://doi.org/10.1038/srep17533.
Khoo BL, Lee SC, Kumar P, Tan TZ, Warkiani ME, Ow SGW, et al. Short-term expansion of breast circulating cancer cells predicts response to anti-cancer therapy. Oncotarget. 2015;6(17):15578–93. https://doi.org/10.18632/oncotarget.3903.
Zhang L, Ridgway LD, Wetzel MD, Ngo J, Yin W, Kumar D, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5(180):180ra48. https://doi.org/10.1126/scitranslmed.3005109.
Cayrefourcq L, Mazard T, Joosse S, Solassol J, Ramos J, Assenat E, et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015;75(5):892–901. https://doi.org/10.1158/0008-5472.CAN-14-2613.
Soler A, Cayrefourcq L, Mazard T, Babayan A, Lamy P-J, Assou S, et al. Autologous cell lines from circulating colon cancer cells captured from sequential liquid biopsies as model to study therapy-driven tumor changes. Sci Rep. 2018;8(1):15931. https://doi.org/10.1038/s41598-018-34365-z.
Zhao P, Zhou W, Liu C, Zhang H, Cheng Z, Wu W, et al. Establishment and characterization of a CTC cell line from peripheral blood of breast cancer patient. J Cancer. 2019;10(24):6095–104. https://doi.org/10.7150/jca.33157.
Dairkee SH, Ji Y, Ben Y, Moore DH, Meng Z, Jeffrey SS. A molecular ‘signature’ of primary breast cancer cultures; patterns resembling tumor tissue. BMC Genomics. 2004;5(1):47. https://doi.org/10.1186/1471-2164-5-47.
Dairkee SH, Nicolau M, Sayeed A, Champion S, Ji Y, Moore DH, et al. Oxidative stress pathways highlighted in tumor cell immortalization: association with breast cancer outcome. Oncogene. 2007;26(43):6269–79. https://doi.org/10.1038/sj.onc.1210452.
Daniel VC, Marchionni L, Hierman JS, Rhodes JT, Devereux WL, Rudin CM, et al. A primary xenograft model of small-cell lung cancer reveals irreversible changes in gene expression imposed by culture in vitro. Cancer Res. 2009;69(8):3364–73. https://doi.org/10.1158/0008-5472.CAN-08-4210.
Maykel J, Liu JH, Li H, Shultz LD, Greiner DL, Houghton J. NOD-scidIl2rg (tm1Wjl) and NOD-Rag1 (null) Il2rg (tm1Wjl) : a model for stromal cell-tumor cell interaction for human colon cancer. Dig Dis Sci. 2014;59(6):1169–79. https://doi.org/10.1007/s10620-014-3168-5.
Zhang H, Cohen AL, Krishnakumar S, Wapnir IL, Veeriah S, Deng G, et al. Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition. Breast Cancer Res. 2014;16(2):R36. https://doi.org/10.1186/bcr3640.
Dobrolecki LE, Airhart SD, Alferez DG, Aparicio S, Behbod F, Bentires-Alj M, et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016;35(4):547–73. https://doi.org/10.1007/s10555-016-9653-x.
Bruna A, Rueda OM, Greenwood W, Batra AS, Callari M, Batra RN, et al. A biobank of breast cancer explants with preserved intra-tumor heterogeneity to screen anticancer compounds. Cell. 2016;167(1):260-74.e22. https://doi.org/10.1016/j.cell.2016.08.041.
DeRose YS, Gligorich KM, Wang G, Georgelas A, Bowman P, Courdy SJ, et al. Patient-derived models of human breast cancer: protocols for in vitro and in vivo applications in tumor biology and translational medicine. Curr Protoc Pharmacol. 2013;Chapter 14:Unit14.23. https://doi.org/10.1002/0471141755.ph1423s60.
Ramani VC, Lemaire CA, Triboulet M, Casey KM, Heirich K, Renier C, et al. Investigating circulating tumor cells and distant metastases in patient-derived orthotopic xenograft models of triple-negative breast cancer. Breast Cancer Res. 2019;21(1):98. https://doi.org/10.1186/s13058-019-1182-4.
Agnoletto C, Minotti L, Brulle-Soumare L, Pasquali L, Galasso M, Corra F, et al. Heterogeneous expression of EPCAM in human circulating tumour cells from patient-derived xenografts. Biomarker Res. 2018;6:31. https://doi.org/10.1186/s40364-018-0145-8.
Lawson DA, Bhakta NR, Kessenbrock K, Prummel KD, Yu Y, Takai K, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–5. https://doi.org/10.1038/nature15260.
Powell E, Shao J, Yuan Y, Chen H-C, Cai S, Echeverria GV, et al. p53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancer. Breast Cancer Res. 2016;18(1):13. https://doi.org/10.1186/s13058-016-0673-9.
Ramirez AB, Bhat R, Sahay D, De Angelis C, Thangavel H, Hedayatpour S, et al. Circulating tumor cell investigation in breast cancer patient-derived xenograft models by automated immunofluorescence staining, image acquisition, and single cell retrieval and analysis. BMC Cancer. 2019;19(1):220. https://doi.org/10.1186/s12885-019-5382-1.
Tachtsidis A, Le AV-P, Blick T, Gunasinghe D, De Sousa E, Waltham M, et al. Human-specific RNA analysis shows uncoupled epithelial-mesenchymal plasticity in circulating and disseminated tumour cells from human breast cancer xenografts. Clin Exp Metastasis. 2019;36(4):393–409. https://doi.org/10.1007/s10585-019-09977-y.
Lallo A, Schenk MW, Frese KK, Blackhall F, Dive C. Circulating tumor cells and CDX models as a tool for preclinical drug development. Transl Lung Cancer Res. 2017;6(4):397–408. https://doi.org/10.21037/tlcr.2017.08.01.
Morrow CJ, Trapani F, Metcalf RL, Bertolini G, Hodgkinson CL, Khandelwal G, et al. Tumourigenic non-small-cell lung cancer mesenchymal circulating tumour cells: a clinical case study. Ann Oncol. 2016;27(6):1155–60. https://doi.org/10.1093/annonc/mdw122.
Que Z, Luo B, Zhou Z, Dong C, Jiang Y, Wang L, et al. Establishment and characterization of a patient-derived circulating lung tumor cell line in vitro and in vivo. Cancer Cell Int. 2019;19:21. https://doi.org/10.1186/s12935-019-0735-z.
Williamson SC, Metcalf RL, Trapani F, Mohan S, Antonello J, Abbott B, et al. Vasculogenic mimicry in small cell lung cancer. Nat Commun. 2016;7:13322. https://doi.org/10.1038/ncomms13322.
Vishnoi M, Boral D, Liu H, Sprouse ML, Yin W, Goswami-Sewell D, et al. Targeting USP7 identifies a metastasis-competent state within bone marrow-resident melanoma CTCs. Cancer Res. 2018;78(18):5349–62. https://doi.org/10.1158/0008-5472.CAN-18-0644.
Pereira-Veiga T, Abreu M, Robledo D, Matias-Guiu X, Santacana M, Sanchez L, et al. CTCs-derived xenograft development in a triple negative breast cancer case. Int J Cancer. 2019;144(9):2254–65. https://doi.org/10.1002/ijc.32001.
Vishnoi M, Liu NH, Yin W, Boral D, Scamardo A, Hong D, et al. The identification of a TNBC liver metastasis gene signature by sequential CTC-xenograft modeling. Mol Oncol. 2019;13(9):1913–26. https://doi.org/10.1002/1878-0261.12533.
Lallo A, Gulati S, Schenk MW, Khandelwal G, Berglund UW, Pateras IS, et al. Ex vivo culture of cells derived from circulating tumour cell xenograft to support small cell lung cancer research and experimental therapeutics. Br J Pharmacol. 2019;176(3):436–50. https://doi.org/10.1111/bph.14542.
Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–44. https://doi.org/10.1038/nbt.2576.
Rossi E, Rugge M, Facchinetti A, Pizzi M, Nardo G, Barbieri V, et al. Retaining the long-survive capacity of Circulating Tumor Cells (CTCs) followed by xeno-transplantation: not only from metastatic cancer of the breast but also of prostate cancer patients. Oncoscience. 2014;1(1):49–56. https://doi.org/10.18632/oncoscience.8.
Ameri K, Luong R, Zhang H, Powell AA, Montgomery KD, Espinosa I, et al. Circulating tumour cells demonstrate an altered response to hypoxia and an aggressive phenotype. Br J Cancer. 2010;102(3):561–9. https://doi.org/10.1038/sj.bjc.6605491.
Das DK, Naidoo MK, Ilboudo A, DuBois P, Durojaiye V, Liu C, et al. Isolation and propagation of circulating tumor cells from a mouse cancer model. J Visual Exp. 2015;(104). https://doi.org/10.3791/52861.
Kang JH, Krause S, Tobin H, Mammoto A, Kanapathipillai M, Ingber DE. A combined micromagnetic-microfluidic device for rapid capture and culture of rare circulating tumor cells. Lab Chip. 2012;12(12):2175–81. https://doi.org/10.1039/c2lc40072c.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–52. https://doi.org/10.1038/35021093.
Russnes HG, Lingjaerde OC, Borresen-Dale A-L, Caldas C. Breast cancer molecular stratification: from intrinsic subtypes to integrative clusters. Am J Pathol. 2017;187(10):2152–62. https://doi.org/10.1016/j.ajpath.2017.04.022.
Newton PK, Mason J, Venkatappa N, Jochelson MS, Hurt B, Nieva J, et al. Spatiotemporal progression of metastatic breast cancer: a Markov chain model highlighting the role of early metastatic sites. NPJ Breast Cancer. 2015;1:15018. https://doi.org/10.1038/npjbcancer.2015.18.
Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, et al. Genomic evolution of breast cancer metastasis and relapse. Cancer Cell. 2017;32(2):169-84.e7. https://doi.org/10.1016/j.ccell.2017.07.005.
Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin Oncol. 2015;12(7):381–94. https://doi.org/10.1038/nrclinonc.2015.73.
Lee JS, Magbanua MJM, Park JW. Circulating tumor cells in breast cancer: applications in personalized medicine. Breast Cancer Res Treat. 2016;160(3):411–24. https://doi.org/10.1007/s10549-016-4014-6.
Agelaki S, Kalykaki A, Markomanolaki H, Papadaki MA, Kallergi G, Hatzidaki D, et al. Efficacy of lapatinib in therapy-resistant HER2-positive circulating tumor cells in metastatic breast cancer. PLoS One. 2015;10(6):e0123683. https://doi.org/10.1371/journal.pone.0123683.
de Kruijff IE, Sieuwerts AM, Onstenk W, Jager A, Hamberg P, de Jongh FE, et al. Androgen receptor expression in circulating tumor cells of patients with metastatic breast cancer. Int J Cancer. 2019;145(4):1083–9. https://doi.org/10.1002/ijc.32209.
Georgoulias V, Bozionelou V, Agelaki S, Perraki M, Apostolaki S, Kallergi G, et al. Trastuzumab decreases the incidence of clinical relapses in patients with early breast cancer presenting chemotherapy-resistant CK-19mRNA-positive circulating tumor cells: results of a randomized phase II study. Ann Oncol. 2012;23(7):1744–50. https://doi.org/10.1093/annonc/mds020.
Reinhardt F, Franken A, Meier-Stiegen F, Driemel C, Stoecklein NH, Fischer JC, et al. Diagnostic leukapheresis enables reliable transcriptomic profiling of single circulating tumor cells to characterize inter-cellular heterogeneity in terms of endocrine resistance. Cancers (Basel). 2019;11(7). pii: E903. https://doi.org/10.3390/cancers11070903.
Li J, Ai Y, Wang L, Bu P, Sharkey CC, Wu Q, et al. Targeted drug delivery to circulating tumor cells via platelet membrane-functionalized particles. Biomaterials. 2016;76:52–65. https://doi.org/10.1016/j.biomaterials.2015.10.046.
Lu Y, Lian S, Ye Y, Yu T, Liang H, Cheng Y, et al. S-Nitrosocaptopril prevents cancer metastasis in vivo by creating the hostile bloodstream microenvironment against circulating tumor cells. Pharmacol Res. 2019;139:535–49. https://doi.org/10.1016/j.phrs.2018.10.020.
Pantel K, Alix-Panabieres C. Functional studies on viable circulating tumor cells. Clin Chem. 2016;62(2):328–34. https://doi.org/10.1373/clinchem.2015.242537.
Khoo BL, Grenci G, **g T, Lim YB, Lee SC, Thiery JP, et al. Liquid biopsy and therapeutic response: circulating tumor cell cultures for evaluation of anticancer treatment. Sci Adv. 2016;2(7):e1600274. https://doi.org/10.1126/sciadv.1600274.
Khoo BL, Grenci G, Lim YB, Lee SC, Han J, Lim CT. Expansion of patient-derived circulating tumor cells from liquid biopsies using a CTC microfluidic culture device. Nat Protoc. 2018;13(1):34–58. https://doi.org/10.1038/nprot.2017.125.
Yoo B, Fuchs BC, Medarova Z. New directions in the study and treatment of metastatic cancer. Front Oncol. 2018;8:258. https://doi.org/10.3389/fonc.2018.00258.
Narkhede AA, Shevde LA, Rao SS. Biomimetic strategies to recapitulate organ specific microenvironments for studying breast cancer metastasis. Int J Cancer. 2017;141(6):1091–109. https://doi.org/10.1002/ijc.30748.
Liu W, Vivian CJ, Brinker AE, Hampton KR, Lianidou E, Welch DR. Microenvironmental influences on metastasis suppressor expression and function during a metastatic cell’s journey. Cancer Microenviron. 2014;7(3):117–31. https://doi.org/10.1007/s12307-014-0148-4.
Tellez-Gabriel M, Cochonneau D, Cadé M, Jubellin C, Heymann MF, Heymann D. Circulating tumor cell-derived pre-clinical models for personalized medicine. Cancers (Basel). 2018;11(1). pii: E19. https://doi.org/10.3390/cancers11010019.
Acknowledgements
The authors wish to thank Dr. Ahmet F. Coskun for his critical reading and comments on this manuscript.
Conflicts of Interest
Dr. Ramalingam is an employee and stockholder of Fluidigm Corporation. The other authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Kujur, P.K., Flores, B.C.T., Ramalingam, N., Chinen, L.T.D., Jeffrey, S.S. (2020). Advances in the Characterization of Circulating Tumor Cells in Metastatic Breast Cancer: Single Cell Analyses and Interactions, and Patient-Derived Models for Drug Testing. In: Piñeiro, R. (eds) Circulating Tumor Cells in Breast Cancer Metastatic Disease. Advances in Experimental Medicine and Biology, vol 1220. Springer, Cham. https://doi.org/10.1007/978-3-030-35805-1_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-35805-1_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-35804-4
Online ISBN: 978-3-030-35805-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)