Abstract
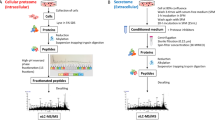
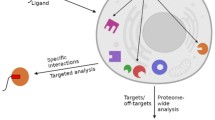
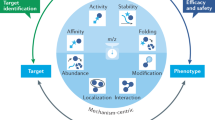
DNA-based technologies such as RNAi, chemical-genetic profiling, or gene expression profiling by DNA microarrays combined with other biochemical methods are established strategies for surveying drug mechanisms. Such approaches can provide mechanistic information on how drugs act and affect cellular pathways. By studying how cancer cells compensate for the drug treatment, novel targets used in a combined treatment can be designed. Furthermore, toxicity effects on cells not targeted can be obtained on a molecular level. For example, drug companies are particularly interested in studying the molecular side effects of drugs in the liver. In addition, experiments with the purpose of elucidating liver toxicity can be studied using samples obtained from animal models exposed to different concentrations of a drug over time. More recently considerable advances in mass spectrometry (MS) technologies and bioinformatics tools allows informative global drug profiling experiments to be performed at a cost comparable to other large-scale technologies such as DNA-based technologies. Moreover, MS-based proteomics provides an additional layer of information on the dynamic regulation of proteins translation and particularly protein degradation. MS-based proteomics approaches combined with other biochemical methods delivers information on regulatory networks, signaling cascades, and metabolic pathways upon drug treatment. Furthermore, MS-based proteomics can provide additional information on single amino acid polymorphisms, protein isoform distribution, posttranslational modifications, and subcellular localization. In this chapter, we will share our experience using MS based proteomics as a pharmacoproteomics strategy to characterize drug mechanisms of action in single drug therapy or in multidrug combination. Finally, the emergence of integrated proteogenomics analysis, such as “The Cancer Genome Atlas” program, opened interesting perspectives to extend this approach to drug target discovery and validation.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Drews J (2003) Stategic trends in the drug industry. Drug Discov Today 8(9):411–420
Yildirim MA et al (2007) Drug-target network. Nat Biotechnol 25(10):1119–1126
Schirle M, Bantscheff M, Kuster B (2012) Mass spectrometry-based proteomics in preclinical drug discovery. Chem Biol 19(1):72–84
Carvalho AS et al (2014) Global mass spectrometry and transcriptomics array based drug profiling provides novel insight into glucosamine induced ER stress. Mol Cell Proteomics 13(12):3294–3307
Hess S (2013) The emerging field of chemo- and pharmacoproteomics. Proteomics Clin Appl 7(1–2):171–180
Kim MS et al (2014) Heterogeneity of pancreatic cancer metastases in a single patient revealed by quantitative proteomics. Mol Cell Proteomics 13(11):2803–2811
Le Bihan T, Duewel HS, Figeys D (2003) On-line strong cation exchange micro-HPLC-ESI-MS/MS for protein identification and process optimization. J Am Soc Mass Spectrom 14(7):719–727
Schirmer EC, Yates JR 3rd, Gerace L (2003) MudPIT: a powerful proteomics tool for discovery. Discov Med 3(18):38–39
Graham JM (2001) Isolation of mitochondria from tissues and cells by differential centrifugation. Curr Protoc Cell Biol Chapter 3. Unit 3.3
Carvalho AS et al (2016) New insights into functional regulation in MS-based drug profiling, Scientific Reports 6:18826
Shevchenko A et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1(6):2856–2860
Manza LL et al (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5(7):1742–1745
Wisniewski JR et al (2009) Universal sample preparation method for proteome analysis. Nat Methods 6(5):359–362
Matthiesen R, Carvalho AS (2013) Methods and algorithms for quantitative proteomics by mass spectrometry. Methods Mol Biol 1007:183–217
Matthiesen R (2013) LC-MS spectra processing. Methods Mol Biol 1007:47–63
Matthiesen R, Carvalho AS (2010) Methods and algorithms for relative quantitative proteomics by mass spectrometry. Methods Mol Biol 593:187–204
Boersema PJ et al (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc 4(4):484–494
Hsu JL et al (2003) Stable-isotope dimethyl labeling for quantitative proteomics. Anal Chem 75(24):6843–6852
Scholz C et al (2015) Acetylation site specificities of lysine deacetylase inhibitors in human cells. Nat Biotechnol 33(4):415–423
Acknowledgement
This work was supported by EXPL/DTP-PIC/0616/2013. RM is sustained by the Fundação para a Ciência e a Tecnologia (FCT) investigator 2012 program. ASC is supported by grant SFRH/BPD/85569/2012 funded by Fundação para a Ciência e Tecnologia. The authors would like to acknowledge networking support by the Proteostasis COST Action (BM1307).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Carvalho, A.S., Matthiesen, R. (2016). Global MS-Based Proteomics Drug Profiling. In: Matthiesen, R. (eds) Proteostasis. Methods in Molecular Biology, vol 1449. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3756-1_31
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3756-1_31
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3754-7
Online ISBN: 978-1-4939-3756-1
eBook Packages: Springer Protocols