Abstract
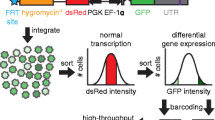
Posttranscriptional control of mRNA subcellular localization, stability, and translation is a central aspect of gene regulation and expression. Much of this control is mediated via recognition of a given mRNA transcript’s 3′ untranslated region (UTR) by microRNAs and RNA-binding proteins. Here we describe how a novel, scalable piggyBac-based vector, pBUTR, can be utilized for analysis of 3′ UTR-mediated posttranscriptional gene regulation (PTGR) both in vitro and in vivo. This vector is specifically designed to express a selection marker, a control reporter, and an experimental reporter from three independent transcription units. Expression of spliced reporter transcripts from medium-copy non-viral promoter elements circumvents several potential confounding factors associated with saturation and stability, while stable integration of these reporter and selection elements in the context of a DNA transposon facilitates experimental reproducibility.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Schena M, Shalon D, Davis RW et al (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470
Cloonan N, Forrest AR, Kolle G et al (2008) Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5:613–619
Mortazavi A, Williams BA, McCue K et al (2008) Map** and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Chen G, Gharib TG, Huang CC et al (2002) Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics 1:304–313
Vogel C, Abreu RS, Ko D et al (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol 6:400
Schwanhausser B, Busse D, Li N et al (2011) Global quantification of mammalian gene expression control. Nature 473:337–342
Jansen RP (2001) mRNA localization: message on the move. Nat Rev Mol Cell Biol 2:247–256
de Moor CH, Meijer H, Lissenden S (2005) Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin Cell Dev Biol 16:49–58
Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8:113–126
Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8:533–543
Merritt C, Rasoloson D, Ko D et al (2008) 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18:1476–1482
Matoulkova E, Michalova E, Vojtesek B et al (2012) The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9:563–576
Kornblihtt AR, Schor IE, Allo M et al (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14:153–165
Elkon R, Ugalde AP, Agami R (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14:496–506
Conne B, Stutz A, Vassalli JD (2000) The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med 6:637–641
Shibayama A, Cook EH Jr, Feng J et al (2004) MECP2 structural and 3′-UTR variants in schizophrenia, autism and other psychiatric diseases: a possible association with autism. Am J Med Genet B Neuropsychiatr Genet 128B:50–53
Chatterjee S, Pal JK (2009) Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol Cell 101:251–262
Chaudhury A, Kongchan N, Gengler JP et al (2014) A piggyBac-based reporter system for scalable in vitro and in vivo analysis of 3′ untranslated region-mediated gene regulation. Nucleic Acids Res 42, e86
Nakano K, Ando T, Yamagishi M et al (2013) Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: implications for retroviral replication. Microbes Infect 15:491–505
Zufferey R, Donello JE, Trono D et al (1999) Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol 73:2886–2892
Jahner D, Stuhlmann H, Stewart CL et al (1982) De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298:623–628
Yoder JA, Walsh CP, Bestor TH (1997) Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13:335–340
Jones S, Peng PD, Yang S et al (2009) Lentiviral vector design for optimal T cell receptor gene expression in the transduction of peripheral blood lymphocytes and tumor-infiltrating lymphocytes. Hum Gene Ther 20:630–640
Hartley JL, Temple GF, Brasch MA (2000) DNA cloning using in vitro site-specific recombination. Genome Res 10:1788–1795
**a H, Mao Q, Davidson BL (2001) The HIV Tat protein transduction domain improves the biodistribution of beta-glucuronidase expressed from recombinant viral vectors. Nat Biotechnol 19:640–644
Gregory PA, Bert AG, Paterson EL et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10:593–601
Yusa K, Zhou L, Li MA et al (2011) A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci U S A 108:1531–1536
Eggan K, Akutsu H, Loring J et al (2001) Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A 98:6209–6214
http://www.hudsonalpha.org/gsl/information/software/bam2fastq.
Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17:438–442
Cary LC, Goebel M, Corsaro BG et al (1989) Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 172:156–169
Li MA, Turner DJ, Ning Z et al (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res 39, e148
Ding S, Wu X, Li G et al (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell 122:473–478
Wu SC, Meir YJ, Coates CJ et al (2006) piggyBac is a flexible and highly active transposon as compared to slee** beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci U S A 103:15008–15013
Nakanishi H, Higuchi Y, Kawakami S et al (2010) piggyBac transposon-mediated long-term gene expression in mice. Mol Ther 18:707–714
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Chaudhury, A., Neilson, J.R. (2016). Use of the pBUTR Reporter System for Scalable Analysis of 3′ UTR-Mediated Gene Regulation. In: Dassi, E. (eds) Post-Transcriptional Gene Regulation. Methods in Molecular Biology, vol 1358. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-3067-8_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3067-8_7
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-3066-1
Online ISBN: 978-1-4939-3067-8
eBook Packages: Springer Protocols