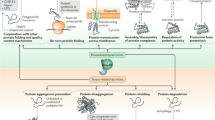
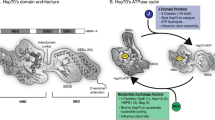
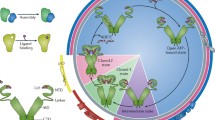
Abstract
The heat shock protein (Hsp)70 protein chaperone is a ubiquitous and promiscuous simple machine that functions in a wide variety of cellular processes. Its primary role of binding short, exposed hydrophobic stretches in misfolded polypeptides is augmented by the participation of an array of partners that act at multiple levels to govern Hsp70 functionality. Protein folding by Hsp70 is adenosine triphosphate (ATP)-dependent and the state of nucleotide binding is driven by dedicated Hsp70 cofactors. Moreover, these and other associates provide functional specificity by virtue of pathway-specific interactions that both recruit and regulate Hsp70 to provide critical protein remodeling contributions. In this chapter, we break down these interactions into two broad themes: protein biogenesis and maturation, and quality control surveillance and degradation. Key findings that establish the pathway- or process-specific network around Hsp70 supporting each cellular activity are discussed, with special attention paid to protein interactions that dictate the contextual role Hsp70 plays. Understanding these various chaperone networks is a requisite first step in designing pathway-specific pharmacological agents for potential therapeutic intervention in protein misfolding disorders.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272(5268):1606–1614
Saibil H (2013) Chaperone machines for protein folding, unfolding and disaggregation. Nat Rev Cancer 13(10):630–642. doi:10.1038/nrm3658
Vogel M, Mayer MP, Bukau B (2006) Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem 281(50):38705–38711. doi:10.1074/jbc.M609020200
Popp S, Packschies L, Radzwill N, Vogel KP, Steinhoff HJ, Reinstein J (2005) Structural dynamics of the DnaK-peptide complex. J Mol Biol 347(5):1039–1052. doi:10.1016/j.jmb.2005.02.026
Flaherty KM, DeLuca-Flaherty C, McKay DB (1990) Three-dimensional structure of the ATPase fragment of a 70 K heat-shock cognate protein. Nature 346(6285):623–628
Takeda S, McKay DB (1996) Kinetics of peptide binding to the bovine 70 kDa heat shock cognate protein, a molecular chaperone. BioChemistry 35(14):4636–4644. doi:10.1021/bi952903o
Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J (1996) The second step of ATP binding to DnaK induces peptide release. J Mol Biol 263(5):657–670. doi:10.1006/jmbi.1996.0606
Schlecht R, Erbse AH, Bukau B, Mayer MP (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol 18(3):345–351. doi:10.1038/nsmb.2006
Kityk R, Kopp J, Sinning I, Mayer MP (2012) Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell 48(6):863–874. doi:10.1016/j.molcel.2012.09.023
Wu CC, Naveen V, Chien CH, Chang YW, Hsiao CD (2012) Crystal structure of DnaK protein complexed with nucleotide exchange factor GrpE in DnaK chaperone system: insight into intermolecular communication. J Biol Chem 287(25):21461–21470. doi:10.1074/jbc.M112.344358
Zhuravleva A, Clerico EM, Gierasch LM (2012) An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell 151(6):1296–1307. doi:10.1016/j.cell.2012.11.002
Davis JE, Voisine C, Craig EA (1999) Intragenic suppressors of Hsp70 mutants: interplay between the ATPase- and peptide-binding domains. Proc Natl Acad Sci U S A 96(16):9269–9276
Craig EA, Huang P, Aron R, Andrew A (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol Biochem Pharmacol 156:1–21
Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell 28(3):422–433. doi:10.1016/j.molcel.2007.08.022
Johnson BD, Schumacher RJ, Ross ED, Toft DO (1998) Hop modulates Hsp70/Hsp90 interactions in protein folding. J Biol Chem 273(6):3679–3686
Tsai J, Douglas MG (1996) A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem 271(16):9347–9354
Cyr DM (1995) Cooperation of the molecular chaperone Ydj1 with specific Hsp70 homologs to suppress protein aggregation. FEBS Lett 359(2–3):129–132
Li J, Qian X, Sha B (2003) The crystal structure of the yeast Hsp40 Ydj1 complexed with its peptide substrate. Structure 11(12):1475–1483
Wall D, Zylicz M, Georgopoulos C (1995) The conserved G/F motif of the DnaJ chaperone is necessary for the activation of the substrate binding properties of the DnaK chaperone. J Biol Chem 270(5):2139–2144
Kam**a HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11 (8):579–592. doi:nrm2941 [pii], 10.1038/nrm2941
Johnson JL, Craig EA (2001) An essential role for the substrate-binding region of Hsp40 s in Saccharomyces cerevisiae. J Cell Biol 152(4):851–856
Liu Q, Hendrickson WA (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131(1):106–120. doi:10.1016/j.cell.2007.08.039
Polier S, Dragovic Z, Hartl FU, Bracher A (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133(6):1068–1079. doi:10.1016/j.cell.2008.05.022
Schuermann JP, Jiang J, Cuellar J, Llorca O, Wang L, Gimenez LE, ** S, Taylor AB, Demeler B, Morano KA, Hart PJ, Valpuesta JM, Lafer EM, Sousa R (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol Cell 31(2):232–243. doi:10.1016/j.molcel.2008.05.006
Trott A, Shaner L, Morano KA (2005) The molecular chaperone Sse1 and the growth control protein kinase Sch9 collaborate to regulate protein kinase A activity in Saccharomyces cerevisiae. Genetics 170(3):1009–1021. doi:10.1534/genetics.105.043109
Makhnevych T, Wong P, Pogoutse O, Vizeacoumar FJ, Greenblatt JF, Emili A, Houry WA (2012) Hsp110 is required for spindle length control. J Cell Biol 198(4):623–636. doi:10.1083/jcb.201111105
Shaner L, Gibney PA, Morano KA (2008) The Hsp110 protein chaperone Sse1 is required for yeast cell wall integrity and morphogenesis. Curr Genet 54(1):1–11. doi:10.1007/s00294-008-0193-y
Oh HJ, Easton D, Murawski M, Kaneko Y, Subjeck JR (1999) The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J Biol Chem 274(22):15712–15718
Park J, Easton DP, Chen X, MacDonald IJ, Wang XY, Subjeck JR (2003) The chaperoning properties of mouse grp170, a member of the third family of hsp70 related proteins. BioChemistry 42(50):14893–14902. doi:10.1021/bi030122e
Anttonen AK, Mahjneh I, Hamalainen RH, Lagier-Tourenne C, Kopra O, Waris L, Anttonen M, Joensuu T, Kalimo H, Paetau A, Tranebjaerg L, Chaigne D, Koenig M, Eeg-Olofsson O, Udd B, Somer M, Somer H, Lehesjoki AE (2005) The gene disrupted in Marinesco-Sjogren syndrome encodes SIL1, an HSPA5 cochaperone. Nat Genet 37(12):1309–1311
Goeckeler JL, Petruso AP, Aguirre J, Clement CC, Chiosis G, Brodsky JL (2008) The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett 582(16):2393–2396. doi:S0014-5793(08)00477-8 [pii], 10.1016/j.febslet.2008.05.047
Polier S, Hartl FU, Bracher A (2010) Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J Mol Biol 401(5):696–707. doi:10.1016/j.jmb.2010.07.004
Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, Liu Q (2012) Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol Chem 287(8):5661–5672. doi:10.1074/jbc.M111.275057
Pechmann S, Willmund F, Frydman J (2013) The ribosome as a hub for protein quality control. Mol Cell 49(3):411–421. doi:10.1016/j.molcel.2013.01.020
Duttler S, Pechmann S, Frydman J (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 50(3):379–393. doi:10.1016/j.molcel.2013.03.010
Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400(6745):693–696. doi:10.1038/23301
Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97(6):755–765
Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B (2003) Trigger factor and DnaK possess overlap** substrate pools and binding specificities. Mol Microbiol 47(5):1317–1328
Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B (2008) Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem 283(7):4124–4132. doi:10.1074/jbc.M708294200
Preissler S, Deuerling E (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37(7):274–283. doi:10.1016/j.tibs.2012.03.002
Huang P, Gautschi M, Walter W, Rospert S, Craig EA (2005) The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol 12(6):497–504
Yam AY, Albanese V, Lin HT, Frydman J (2005) Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J Biol Chem 280(50):41252–41261. doi:10.1074/jbc.M503615200
Hundley HA, Walter W, Bairstow S, Craig EA (2005) Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science 308(5724):1032–1034
Jaiswal H, Conz C, Otto H, Wolfle T, Fitzke E, Mayer MP, Rospert S (2011) The chaperone network connected to human ribosome-associated complex. Mol Cell Biol 31(6):1160–1173. doi:10.1128/MCB.00986-10
Broadley SA, Hartl FU (2009) The role of molecular chaperones in human misfolding diseases. FEBS Lett 583(16):2647–2653. doi:10.1016/j.febslet.2009.04.029
Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA (1998) The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J 17(14):3981–3989. doi:10.1093/emboj/17.14.3981
James P, Pfund C, Craig EA (1997) Functional specificity among Hsp70 molecular chaperones. Science 275(5298):387–389
Peisker K, Chiabudini M, Rospert S (2010) The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim Biophys Acta 1803(6):662–672. doi:10.1016/j.bbamcr.2010.03.005
Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I (2013) Structural characterization of a eukaryotic chaperone-the ribosome-associated complex. Nat Struct Mol Biol 20(1):23–28. doi:10.1038/nsmb.2447
Fiaux J, Horst J, Scior A, Preissler S, Koplin A, Bukau B, Deuerling E (2010) Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J Biol Chem 285(5):3227–3234. doi:M109.075804 [pii], 10.1074/jbc.M109.075804
Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU (2012) DnaK functions as a central hub in the E. coli chaperone network. Cell Rep 1 (3):251–264. doi:10.1016/j.celrep. 2011.12.007
Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA (1998) Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 17(16):4809–4817. doi:10.1093/emboj/17.16.4809
Peisker K, Braun D, Wolfle T, Hentschel J, Funfschilling U, Fischer G, Sickmann A, Rospert S (2008) Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell 19(12):5279–5288. doi:10.1091/mbc.E08- 06-0661
Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, Houry WA (2009) An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol 5:275. doi:msb200926 [pii], 10.1038/msb.2009.26
Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J (2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152(1-2):196–209. doi:10.1016/j.cell.2012.12.001
Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S (2005) The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A 102(29):10064–10069. doi:10.1073/pnas.0504400102
Shaner L, Sousa R, Morano KA (2006) Characterization of Hsp70 binding and nucleotide exchange by the yeast Hsp110 chaperone Sse1. BioChemistry 45(50):15075–15084. doi:10.1021/bi061279k
Oh HJ, Chen X, Subjeck JR (1997) Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J Biol Chem 272(50):31636–31640
Verghese J, Morano KA (2012) A lysine-rich region within fungal BAG domain-containing proteins mediates a novel association with ribosomes. Eukaryot Cell 11(8):1003–1011. doi:10.1128/EC.00146-12
Kurzchalia TV, Wiedmann M, Girshovich AS, Bochkareva ES, Bielka H, Rapoport TA (1986) The signal sequence of nascent preprolactin interacts with the 54 K polypeptide of the signal recognition particle. Nature 320(6063):634–636. doi:10.1038/320634a0
Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R (2004) Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427(6977):808–814. doi:10.1038/nature02342
Walter P, Blobel G (1981) Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol 91(2 Pt 1):557–561
Becker J, Walter W, Yan W, Craig EA (1996) Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol Cell Biol 16(8):4378–4386
Karbstein K (2010) Chaperoning ribosome assembly. J Cell Biol 189(1):11–12. doi:10.1083/jcb.201002102
Albanese V, Reissmann S, Frydman J (2010) A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189(1):69–81. doi:10.1083/jcb.201001054
Meyer AE, Hung NJ, Yang P, Johnson AW, Craig EA (2007) The specialized cytosolic J-protein, Jjj1, functions in 60 S ribosomal subunit biogenesis. Proc Natl Acad Sci U S A 104(5):1558–1563. doi:10.1073/pnas.0610704104
Meyer AE, Hoover LA, Craig EA (2010) The cytosolic J-protein, Jjj1, and Rei1 function in the removal of the pre-60 S subunit factor Arx1. J Biol Chem 285(2):961–968. doi:10.1074/jbc.M109.038349
Herrmann JM, Stuart RA, Craig EA, Neupert W (1994) Mitochondrial heat shock protein 70, a molecular chaperone for proteins encoded by mitochondrial DNA. J Cell Biol 127(4):893–902
Maki JA, Southworth DR, Culver GM (2003) Demonstration of the role of the DnaK chaperone system in assembly of 30 S ribosomal subunits using a purified in vitro system. RNA 9(12):1418–1421
Maki JA, Schnobrich DJ, Culver GM (2002) The DnaK chaperone system facilitates 30S ribosomal subunit assembly. Mol Cell 10(1):129–138
Alix JH, Guerin MF (1993) Mutant DnaK chaperones cause ribosome assembly defects in Escherichia coli. Proc Natl Acad Sci U S A 90(20):9725–9729
Martinez-Hackert E, Hendrickson WA (2009) Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138(5):923–934. doi:10.1016/j.cell.2009.07.044
Meunier L, Usherwood YK, Chung KT, Hendershot LM (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell 13(12):4456–4469. doi:10.1091/mbc.E02-05-0311
Brodsky JL, Werner ED, Dubas ME, Goeckeler JL, Kruse KB, McCracken AA (1999) The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J Biol Chem 274(6):3453–3460
Steel GJ, Fullerton DM, Tyson JR, Stirling CJ (2004) Coordinated activation of Hsp70 chaperones. Science 303(5654):98–101. doi:10.1126/science.1092287
Weitzmann A, Baldes C, Dudek J, Zimmermann R (2007) The heat shock protein 70 molecular chaperone network in the pancreatic endoplasmic reticulum—a quantitative approach. FEBS J 274(19):5175–5187. doi:10.1111/j.1742-4658.2007.06039.x
Hale SJ, Lovell SC, de Keyzer J, Stirling CJ (2010) Interactions between Kar2p and its nucleotide exchange factors Sil1p and Lhs1p are mechanistically distinct. J Biol Chem 285(28):21600–21606. doi:10.1074/jbc.M110.111211
Simons JF, Ferro-Novick S, Rose MD, Helenius A (1995) BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol 130(1):41–49
Lin HY, Masso-Welch P, Di YP, Cai JW, Shen JW, Subjeck JR (1993) The 170-kDa glucose-regulated stress protein is an endoplasmic reticulum protein that binds immunoglobulin. Mol Biol Cell 4(11):1109–1119
Craven RA, Egerton M, Stirling CJ (1996) A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. Embo J 15(11):2640–2650
Senderek J, Krieger M, Stendel C, Bergmann C, Moser M, Breitbach-Faller N, Rudnik-Schoneborn S, Blaschek A, Wolf NI, Harting I, North K, Smith J, Muntoni F, Brockington M, Quijano-Roy S, Renault F, Herrmann R, Hendershot LM, Schroder JM, Lochmuller H, Topaloglu H, Voit T, Weis J, Ebinger F, Zerres K (2005) Mutations in SIL1 cause Marinesco-Sjogren syndrome, a cerebellar ataxia with cataract and myopathy. Nat Genet 37(12):1312–1314. doi:10.1038/ng1678
Tyson JR, Stirling CJ (2000) LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. Embo J 19(23):6440–6452. doi:10.1093/emboj/19.23.6440
Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K (2008) ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science 321(5888):569–572. doi:10.1126/science.1159293
Hagiwara M, Maegawa K, Suzuki M, Ushioda R, Araki K, Matsumoto Y, Hoseki J, Nagata K, Inaba K (2011) Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol Cell 41(4):432–444. doi:10.1016/j.molcel.2011.01.021
Mizzen LA, Chang C, Garrels JI, Welch WJ (1989) Identification, characterization, and purification of two mammalian stress proteins present in mitochondria, grp 75, a member of the hsp 70 family and hsp 58, a homolog of the bacterial groEL protein. J Biol Chem 264(34):20664–20675
Bhattacharyya T, Karnezis AN, Murphy SP, Hoang T, Freeman BC, Phillips B, Morimoto RI (1995) Cloning and subcellular localization of human mitochondrial hsp70. J Biol Chem 270(4):1705–1710
Syken J, De-Medina T, Munger K (1999) TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci U S A 96(15):8499–8504
Voos W, Rottgers K (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta 1592(1):51–62
Voos W, Gambill BD, Laloraya S, Ang D, Craig EA, Pfanner N (1994) Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol Cell Biol 14(10):6627–6634
Andrew AJ, Dutkiewicz R, Knieszner H, Craig EA, Marszalek J (2006) Characterization of the interaction between the J-protein Jac1p and the scaffold for Fe-S cluster biogenesis, Isu1p. J Biol Chem 281(21):14580–14587. doi:10.1074/jbc.M600842200
Dutkiewicz R, Schilke B, Knieszner H, Walter W, Craig EA, Marszalek J (2003) Ssq1, a mitochondrial Hsp70 involved in iron-sulfur (Fe/S) center biogenesis. Similarities to and differences from its bacterial counterpart. J Biol Chem 278(32):29719–29727
Uzarska MA, Dutkiewicz R, Freibert SA, Lill R, Muhlenhoff U (2013) The mitochondrial Hsp70 chaperone Ssq1 facilitates Fe/S cluster transfer from Isu1 to Grx5 by complex formation. Mol Biol Cell 24(12):1830–1841. doi:10.1091/mbc.E12-09-0644
Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE (2008) Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. BioChemistry 47(48):12795–12801. doi:10.1021/bi801565j
Chen S, Smith DF (1998) Hop as an adaptor in the heat shock protein 70 (Hsp70) and hsp90 chaperone machinery. J Biol Chem 273(52):35194–35200
Smith DF (2004) Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9(2):109–121
Siligardi G, Panaretou B, Meyer P, Singh S, Woolfson DN, Piper PW, Pearl LH, Prodromou C (2002) Regulation of Hsp90 ATPase activity by the co-chaperone Cdc37p/p50cdc37. J Biol Chem 277(23):20151–20159. doi:10.1074/jbc.M201287200
Heldens L, Hensen SM, Onnekink C, van Genesen ST, Dirks RP, Lubsen NH (2011) An atypical unfolded protein response in heat shocked cells. PLoS ONE 6(8):e23512. doi:10.1371/journal.pone.0023512
Hodson S, Marshall JJ, Burston SG (2012) Map** the road to recovery: the ClpB/Hsp104 molecular chaperone. J Struct Biol 179(2):161–171. doi:10.1016/j.jsb.2012.05.015
Werner-Washburne M, Becker J, Kosic-Smithers J, Craig EA (1989) Yeast Hsp70 RNA levels vary in response to the physiological status of the cell. J Bacteriol 171(5):2680–2688
Boorstein WR, Craig EA (1990) Structure and regulation of the SSA4 HSP70 gene of Saccharomyces cerevisiae. J Biol Chem 265(31):18912–18921
Lu Z, Cyr DM (1998) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273(43):27824–27830
Winkler J, Tyedmers J, Bukau B, Mogk A (2012) Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol 198(3):387–404. doi:10.1083/jcb.201201074
Schmitt M, Neupert W, Langer T (1996) The molecular chaperone Hsp78 confers compartment-specific thermotolerance to mitochondria. J Cell Biol 134(6):1375–1386
Lum R, Tkach JM, Vierling E, Glover JR (2004) Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J Biol Chem 279(28):29139–29146. doi:10.1074/jbc.M403777200
Shorter J (2011) The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS ONE 6(10):e26319. doi:10.1371/journal.pone.0026319
Lee J, Kim JH, Biter AB, Sielaff B, Lee S, Tsai FT (2013) Heat shock protein (Hsp) 70 is an activator of the Hsp104 motor. Proc Natl Acad Sci U S A 110(21):8513–8518. doi:10.1073/pnas.1217988110
Haslberger T, Weibezahn J, Zahn R, Lee S, Tsai FT, Bukau B, Mogk A (2007) M domains couple the ClpB threading motor with the DnaK chaperone activity. Mol Cell 25(2):247–260. doi:10.1016/j.molcel.2006.11.008
Desantis ME, Shorter J (2012) The elusive middle domain of Hsp104 and ClpB: location and function. Biochim Biophys Acta 1823(1):29–39. doi:10.1016/j.bbamcr.2011.07.014
Miot M, Reidy M, Doyle SM, Hoskins JR, Johnston DM, Genest O, Vitery MC, Masison DC, Wickner S (2011) Species-specific collaboration of heat shock proteins (Hsp) 70 and 100 in thermotolerance and protein disaggregation. Proc Natl Acad Sci U S A 108(17):6915–6920. doi:10.1073/pnas.1102828108
Seyffer F, Kummer E, Oguchi Y, Winkler J, Kumar M, Zahn R, Sourjik V, Bukau B, Mogk A (2012) Hsp70 proteins bind Hsp100 regulatory M domains to activate AAA+ disaggregase at aggregate surfaces. Nat Struct Mol Biol 19(12):1347–1355. doi:10.1038/nsmb.2442
Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE (2013) Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339(6123):1080–1083. doi:10.1126/science.1233066
Zietkiewicz S, Krzewska J, Liberek K (2004) Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem 279(43):44376–44383. doi:10.1074/jbc.M402405200
Rampelt H, Kirstein-Miles J, Nillegoda NB, Chi K, Scholz SR, Morimoto RI, Bukau B (2012) Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J 31(21):4221–4235. doi:10.1038/emboj.2012.264
Jakob U, Gaestel M, Engel K, Buchner J (1993) Small heat shock proteins are molecular chaperones. J Biol Chem 268(3):1517–1520
Haslbeck M, Braun N, Stromer T, Richter B, Model N, Weinkauf S, Buchner J (2004) Hsp42 is the general small heat shock protein in the cytosol of Saccharomyces cerevisiae. EMBO J 23(3):638–649. doi:10.1038/sj.emboj.7600080, 7600080 [pii]
Cashikar AG, Duennwald M, Lindquist SL (2005) A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem 280 (25):23869–23875. doi:M502854200 [pii], 10.1074/jbc.M502854200
Specht S, Miller SB, Mogk A, Bukau B (2011) Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae. J Cell Biol 195(4):617–629. doi:10.1083/jcb.201106037
Lee GJ, Vierling E (2000) A small heat shock protein cooperates with heat shock protein 70 systems to reactivate a heat-denatured protein. Plant Physiol 122(1):189–198
Garrido C, Paul C, Seigneuric R, Kam**a HH (2012) The small heat shock proteins family: the long forgotten chaperones. Int J Biochem Cell Biol 44(10):1588–1592. doi:10.1016/j.biocel.2012.02.022
Mogk A, Schlieker C, Friedrich KL, Schonfeld HJ, Vierling E, Bukau B (2003) Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem 278(33):31033–31042. doi:10.1074/jbc.M303587200
Goldbaum O, Riedel M, Stahnke T, Richter-Landsberg C (2009) The small heat shock protein HSP25 protects astrocytes against stress induced by proteasomal inhibition. Glia 57(14):1566–1577. doi:10.1002/glia.20870
Shiber A, Breuer W, Brandeis M, Ravid T (2013) Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol Biol Cell 24(13):2076–2087. doi:10.1091/mbc.E13 - 01-0010
Malinovska L, Kroschwald S, Munder MC, Richter D, Alberti S (2012) Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol Biol Cell 23(16):3041–3056. doi:10.1091/mbc.E12- 03-0194
Dokladny K, Zuhl MN, Mandell M, Bhattacharya D, Schneider S, Deretic V, Moseley PL (2013) Regulatory coordination between two major intracellular homeostatic systems: heat shock response and autophagy. J Biol Chem 288(21):14959–14972. doi:10.1074/jbc.M113.462408
Arndt V, Rogon C, Hohfeld J (2007) To be, or not to be–molecular chaperones in protein degradation. Cell Mol Life Sci 64(19-20):2525–2541. doi:10.1007/s00018-007-7188-6
Kravtsova-Ivantsiv Y, Ciechanover A (2012) Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci 125(Pt 3):539–548. doi:10.1242/jcs.093567
Voges D, Zwickl P, Baumeister W (1999) The 26 S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem 68:1015–1068. doi:10.1146/annurev.biochem.68:1.1015
Guerriero CJ, Weiberth KF, Brodsky JL (2013) Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J Biol Chem 288(25):18506–18520. doi:10.1074/jbc.M113.475905
Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, Patterson C (1999) Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 19(6):4535–4545
Buchberger A, Bukau B, Sommer T (2010) Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 40(2):238–252. doi:10.1016/j.molcel.2010.10.001
Murata S, Minami Y, Minami M, Chiba T, Tanaka K (2001) CHIP is a chaperone-dependent E3 ligase that ubiquitylates unfolded protein. EMBO Rep 2(12):1133–1138. doi:10.1093/embo-reports/kve246
Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM (2001) The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol 3(1):100–105. doi:10.1038/35050509
Demand J, Alberti S, Patterson C, Hohfeld J (2001) Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr Biol 11(20):1569–1577
Stankiewicz M, Nikolay R, Rybin V, Mayer MP (2010) CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J 277(16):3353–3367. doi:10.1111/j.1742-4658.2010.07737.x
Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME (2005) HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol 15(11):1058–1064. doi:10.1016/j.cub.2005.04.058
Park SH, Kukushkin Y, Gupta R, Chen T, Konagai A, Hipp MS, Hayer-Hartl M, Hartl FU (2013) PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 154(1):134–145. doi:10.1016/j.cell.2013.06.003
Luders J, Demand J, Hohfeld J (2000) The ubiquitin-related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem 275(7):4613–4617
Kikukawa Y, Minami R, Shimada M, Kobayashi M, Tanaka K, Yokosawa H, Kawahara H (2005) Unique proteasome subunit Xrpn10c is a specific receptor for the antiapoptotic ubiquitin-like protein Scythe. FEBS J 272(24):6373–6386. doi:10.1111/j.1742-4658.2005.05032.x
Gowda NK, Kandasamy G, Froehlich MS, Dohmen RJ, Andreasson C (2013) Hsp70 nucleotide exchange factor Fes1 is essential for ubiquitin-dependent degradation of misfolded cytosolic proteins. Proc Natl Acad Sci U S A 110(15):5975–5980. doi:10.1073/pnas.1216778110
Arndt V, Daniel C, Nastainczyk W, Alberti S, Hohfeld J (2005) BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol Biol Cell 16(12):5891–5900. doi:10.1091/mbc.E05-07-0660
Gebauer M, Zeiner M, Gehring U (1997) Proteins interacting with the molecular chaperone hsp70/hsc70: physical associations and effects on refolding activity. FEBS Lett 417(1):109–113
Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C (2006) CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440(7083):551–555. doi:10.1038/nature04600
Alberti S, Demand J, Esser C, Emmerich N, Schild H, Hohfeld J (2002) Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J Biol Chem 277(48):45920–45927. doi:10.1074/jbc.M204196200
Denic V, Quan EM, Weissman JS (2006) A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 126(2):349–359. doi:10.1016/j.cell.2006.05.045
Kabani M, Kelley SS, Morrow MW, Montgomery DL, Sivendran R, Rose MD, Gierasch LM, Brodsky JL (2003) Dependence of endoplasmic reticulum-associated degradation on the peptide binding domain and concentration of BiP. Mol Biol Cell 14(8):3437–3448. doi:10.1091/mbc.E02-12-0847
Soroka J, Wandinger SK, Mausbacher N, Schreiber T, Richter K, Daub H, Buchner J (2012) Conformational switching of the molecular chaperone Hsp90 via regulated phosphorylation. Mol Cell 45(4):517–528. doi:10.1016/j.molcel.2011.12.031
Truman AW, Kristjansdottir K, Wolfgeher D, Hasin N, Polier S, Zhang H, Perrett S, Prodromou C, Jones GW, Kron SJ (2012) CDK-dependent Hsp70 Phosphorylation controls G1 cyclin abundance and cell-cycle progression. Cell 151(6):1308–1318. doi:10.1016/j.cell.2012.10.051
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Garcia, V., Morano, K. (2014). The Chaperone Networks: A Heat Shock Protein (Hsp)70 Perspective. In: Houry, W. (eds) The Molecular Chaperones Interaction Networks in Protein Folding and Degradation. Interactomics and Systems Biology, vol 1. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1130-1_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1130-1_4
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1129-5
Online ISBN: 978-1-4939-1130-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)