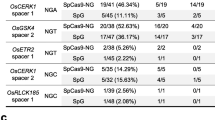
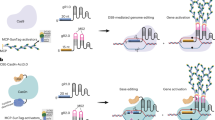
Abstract
Engineered SpCas9 variant, SpRY, has been demonstrated to facilitate protospacer adjacent motif (PAM) unrestricted targeting of genomic DNA in various biological systems. Here we describe fast, efficient, and robust preparation of SpRY-derived genome and base editors that can be easily adapted to target various DNA sequences in plants due to modular Gateway assembly. Presented are detailed protocols for preparing T-DNA vectors for genome and base editors and for assessing genome editing efficiency through transient expression of these reagents in rice protoplasts.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
**ek M, Chylinski K, Fonfara I et al (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Zetsche B, Gootenberg JS, Abudayyeh OO et al (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. https://doi.org/10.1016/j.cell.2015.09.038
Kim YG, Cha J, Chandrasegaran S (1996) Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93:1156–1160. https://doi.org/10.1073/pnas.93.3.1156
Boch J, Bonas U (2010) Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol 48:419–436. https://doi.org/10.1146/annurev-phyto-080508-081936
Miller JC, Tan S, Qiao G et al (2011) A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 29:143–148. https://doi.org/10.1038/nbt.1755
Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4:639–648. https://doi.org/10.1016/j.dnarep.2004.12.005
Sfeir A, Symington LS (2015) Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714. https://doi.org/10.1016/j.tibs.2015.08.006
Liang F, Han M, Romanienko PJ et al (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci 95:5172–5177. https://doi.org/10.1073/pnas.95.9.5172
Jeggo PA (1998) DNA breakage and repair. Adv Genet 38:185–218. https://doi.org/10.1016/s0065-2660(08)60144-3
Kosicki M, Tomberg K, Bradley A (2018) Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol 36:765–771. https://doi.org/10.1038/nbt.4192
Pickar-Oliver A, Gersbach CA (2019) The next generation of CRISPR–Cas technologies and applications. Nat Rev Mol Cell Biol 20:490–507. https://doi.org/10.1038/s41580-019-0131-5
Friedland AE, Baral R, Singhal P et al (2015) Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16:257. https://doi.org/10.1186/s13059-015-0817-8
Walton RT, Christie KA, Whittaker MN et al (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368:290–296. https://doi.org/10.1126/science.aba8853
Ren Q, Sretenovic S, Liu S et al (2021) PAM-less plant genome editing using a CRISPR–SpRY toolbox. Nat Plants 7:25–33. https://doi.org/10.1038/s41477-020-00827-4
Gaudelli NM, Komor AC, Rees HA et al (2017) Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage. Nature 551:464–471. https://doi.org/10.1038/nature24644
Nishida K, Arazoe T, Yachie N et al (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353(6305):aaf8729. https://doi.org/10.1126/science.aaf8729
Komor AC, Kim YB, Packer MS et al (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424. https://doi.org/10.1038/nature17946
Zhao D, Li J, Li S et al (2021) Glycosylase base editors enable C-to-A and C-to-G base changes. Nat Biotechnol 39(1):35–40. https://doi.org/10.1038/s41587-020-0592-2
Chen L, Park JE, Paa P et al (2020) Precise and programmable C:G to G:C base editing in genomic DNA. bioRxiv:2020.07.21.213827. https://doi.org/10.1101/2020.07.21.213827
Kurt IC, Zhou R, Iyer S et al (2021) CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol 39:41–46. https://doi.org/10.1038/s41587-020-0609-x
Kuscu C, Adli M (2016) CRISPR-Cas9-AID base editor is a powerful gain-of-function screening tool. Nat Methods 13:983–984. https://doi.org/10.1038/nmeth.4076
Wang X, Li J, Wang Y et al (2018) Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat Biotechnol 36:946–949. https://doi.org/10.1038/nbt.4198
Richter MF, Zhao KT, Eton E et al (2020) Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat Biotechnol 38(7):883–891. https://doi.org/10.1038/s41587-020-0453-z
Grünewald J, Zhou R, Garcia SP et al (2019) Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569:433. https://doi.org/10.1038/s41586-019-1161-z
Doman JL, Raguram A, Newby GA et al (2020) Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat Biotechnol 38:620–628. https://doi.org/10.1038/s41587-020-0414-6
Xu Z, Kuang Y, Ren B et al (2021) SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol 22:6. https://doi.org/10.1186/s13059-020-02231-9
Zhang C, Wang Y, Wang F et al (2021) Expanding base editing scope to near-PAMless with engineered CRISPR/Cas9 variants in plants. Mol Plant 14:191–194. https://doi.org/10.1016/j.molp.2020.12.016
Chen S, Liu Z, Lai L et al (2022) Efficient C-to-G base editing with improved target compatibility using engineered deaminase–nCas9 fusions. CRISPR J 5(3):389–396. https://doi.org/10.1089/crispr.2021.0124
Sretenovic S, Liu S, Li G et al (2021) Exploring C-To-G base editing in rice, tomato, and poplar. Front Genome Ed 3:756766. https://doi.org/10.3389/fgeed.2021.756766
Park J, Bae S, Kim J-S (2015) Cas-designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31(24):4014–4016. https://doi.org/10.1093/bioinformatics/btv537
Naito Y, Hino K, Bono H et al (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123. https://doi.org/10.1093/bioinformatics/btu743
Wu Y, He Y, Sretenovic S et al (2022) CRISPR-BETS: a base-editing design tool for generating stop codons. Plant Biotechnol J 20:499–510. https://doi.org/10.1111/pbi.13732
Acknowledgments
This work was supported by the NSF Plant Genome Research Program (award no. IOS-2029889 and IOS-2132693) and the USDA-NIFA Biotechnology Risk Assessment Research Grants Program (award no. 2018-33522-28789) to Y.Q. It was also supported by the National Transgenic Major Project (award no. 2018ZX08020-003), the Technology Innovation and Application Development Program of Chongqing (award no. CSTC2021JSCX-CYLHX0001), and the Natural Science Foundation of Sichuan Province (award no. 2022NSFSC0143) to X.T. and Y.Z.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Sretenovic, S., Tang, X., Ren, Q., Zhang, Y., Qi, Y. (2023). PAM-Less CRISPR-SpRY Genome Editing in Plants. In: Yang, B., Harwood, W., Que, Q. (eds) Plant Genome Engineering. Methods in Molecular Biology, vol 2653. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3131-7_1
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3131-7_1
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3130-0
Online ISBN: 978-1-0716-3131-7
eBook Packages: Springer Protocols