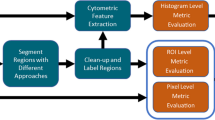
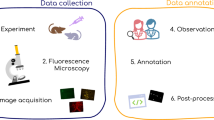
Abstract
The development of automated quantitative image analysis pipelines requires thoughtful considerations to extract meaningful information. Commonly, extraction rules for quantitative parameters are defined and agreed beforehand to ensure repeatability between annotators. Machine/Deep Learning (ML/DL) now provides tools to automatically extract the set of rules to obtain quantitative information from the images (e.g. segmentation, enumeration, classification, etc.). Many parameters must be considered in the development of proper ML/DL pipelines. We herein present the important vocabulary, the necessary steps to create a thorough image segmentation pipeline, and also discuss technical aspects that should be considered in the development of automated image analysis pipelines through ML/DL.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Allan C, Burel JM, Moore J et al (2012) OMERO: flexible, model-driven data management for experimental biology. Nat Methods 9(3):245–253
Alpaydin E (2009) Introduction to machine learning. MIT Press, Cambridge, MA
Anderson J, Mohammed S, Grimm B et al (2011) The Viking viewer for connectomics: scalable multi-user annotation and summarization of large volume data sets. J Microsc 241(1):13–28
Arganda-Carreras I, Kaynig V, Rueden C et al (2017) Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33(15):2424–2426
Berg S, Kutra D, Kroeger T et al (2019) ilastik: interactive machine learning for (bio)image analysis. Nat Methods 1–7
Bilodeau A, Delmas C, Parent M et al (2021) MICRA-Net: MICRoscopy Analysis Neural Network to solve detection, classification, and segmentation from a single simple auxiliary task. bioRxiv 2021.06.29.448970
Boykov Y, Kolmogorov V (2004) An experimental comparison of min-cut/max-flow algorithms for energy minimization in vision. IEEE Trans Pattern Anal Mach Intell 26:1124–1137
Breiman L (2001) Random forests. Mach Learn 45(1):5–32
Caicedo JC, Goodman A, Karhohs KW et al (2019a) Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat Methods 16(12):1247–1253
Caicedo JC, Roth J, Goodman A et al (2019b) Evaluation of deep learning strategies for nucleus segmentation in fluorescence images. Cytom Part A 95(9):952–965
Cordts M, Omran M, Ramos S et al (2016) The cityscapes dataset for semantic urban scene understanding. ar**v:160401685 [cs] 1604.01685
Cortes C, Mohri M, Rostamizadeh A (2012) L2 regularization for learning kernels. ar**v preprint ar**v:12052653
Falk T, Mai D, Bensch R et al (2019) U-net: deep learning for cell counting, detection, and morphometry. Nat Methods 16(1):67
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press. http://www.deeplearningbook.org
Gupta A, Harrison PJ, Wieslander H et al (2019) Deep learning in image cytometry: a review. Cytom Part A 95(4):366–380
He K, Gkioxari G, Dollár P et al (2018) Mask R-CNN. ar**v:170306870 [cs] 1703.06870
Hossin M, Sulaiman M (2015) A review on evaluation metrics for data classification evaluations. Int J Data Min Knowl Manag Process 5(2):1
Ioffe S, Szegedy C (2015) Batch normalization: accelerating deep network training by reducing internal covariate shift. ar**v:150203167 [cs] 1502.03167
Kawaguchi K, Kaelbling LP, Bengio Y (2017) Generalization in deep learning. ar**v preprint ar**v:171005468
Khoreva A, Benenson R, Hosang J et al (2017) Simple does it: weakly supervised instance and semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 876–885
Kiefer J, Wolfowitz J et al (1952) Stochastic estimation of the maximum of a regression function. Ann Math Stat 23(3):462–466
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. ar**v preprint ar**v:14126980
Kotsiantis S, Kanellopoulos D, Pintelas P et al (2006) Handling imbalanced datasets: a review. GESTS Int Trans Comput Sci Eng 30(1):25–36
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Kromp F, Fischer L, Bozsaky E et al (2019) Deep learning architectures for generalized immunofluorescence based nuclear image segmentation. ar**v:190712975 [cs, q-bio] 1907.12975
Kromp F, Bozsaky E, Rifatbegovic F et al (2020) An annotated fluorescence image dataset for training nuclear segmentation methods. Sci Data 7(1):262
Lavoie-Cardinal F, Bilodeau A, Lemieux M et al (2020) Neuronal activity remodels the F-actin based submembrane lattice in dendrites but not axons of hippocampal neurons. Sci Rep 10(1):11960
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436
Lin TY, Maire M, Belongie S et al (2014) Microsoft coco: common objects in context. In: European conference on computer vision. Springer, pp 740–755
Linkert M, Rueden CT, Allan C et al (2010) Metadata matters: access to image data in the real world. J Cell Biol 189(5):777–782
Long J, Shelhamer E, Darrell T (2015) Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 3431–3440
Moen E, Bannon D, Kudo T et al (2019) Deep learning for cellular image analysis. Nature Methods 1–14
Nair V, Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines. In: ICML
Olah C, Mordvintsev A, Schubert L (2017) Feature visualization. Distill https://distill.pub/2017/feature-visualization
Papandreou G, Chen LC, Murphy KP et al (2015) Weakly-and semi-supervised learning of a deep convolutional network for semantic image segmentation. In: Proceedings of the IEEE international conference on computer vision, pp 1742–1750
Qian N (1999) On the momentum term in gradient descent learning algorithms. Neural Netw 12(1):145–151
Raghu M, Zhang C, Kleinberg J et al (2019) Transfusion: understanding transfer learning for medical imaging. In: Wallach H, Larochelle H, Beygelzimer A, d’Alché-Buc F, Fox E, Garnett R (eds) Advances in neural information processing systems, vol 32. Curran Associates, Inc., pp 3347–3357
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Rübel O, Tritt A, Dichter B et al (2019) NWB:N 2.0: an accessible data standard for neurophysiology. bioRxiv p 523035
Sarkans U, Chiu W, Collinson L et al (2021) REMBI: Recommended Metadata for Biological Images—enabling reuse of microscopy data in biology. Nat Methods 1–5
Schermelleh L, Ferrand A, Huser T et al (2019) Super-resolution microscopy demystified. Nat Cell Biol 21(1):72
Schindelin J, Arganda-Carreras I, Frise E et al (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. ar**v preprint ar**v:14091556
Sommer C, Gerlich DW (2013) Machine learning in cell biology–teaching computers to recognize phenotypes. J Cell Sci 126(24):5529–5539
Sommer C, Straehle C, Koethe U et al (2011) Ilastik: interactive learning and segmentation toolkit. In: 2011 IEEE international symposium on biomedical imaging: from nano to macro. IEEE, pp 230–233
Stringer C, Wang T, Michaelos M et al (2021) Cellpose: a generalist algorithm for cellular segmentation. Nat Methods 18(1):100–106
Ulman V, Maška M, Magnusson KE, Ronneberger O, Haubold C, Harder N, Matula P, Matula, P, Svoboda D, Radojevic M, Smal I (2017) An objective comparison of cell tracking algorithms. Nat Methods 14(12):1141–1152
Vicar T, Balvan J, Jaros J others (2019) Cell segmentation methods for label-free contrast microscopy: review and comprehensive comparison. BMC Bioinformatics 20(1):360
Vohs KD, Baumeister RF, Schmeichel BJ et al (2008) Making choices impairs subsequent self-control: a limited-resource account of decision making, self-regulation, and active initiative. J Pers Soc Psychol 94(5):883–898
**e Y, **ng F, Kong X et al (2015) Beyond classification: structured regression for robust cell detection using convolutional neural network. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 358–365
Yang L, Zhang Y, Zhao Z et al (2018) BoxNet: deep learning based biomedical image segmentation using boxes only annotation. ar**v preprint ar**v:180600593
Yeghiazaryan V, Voiculescu ID (2018) Family of boundary overlap metrics for the evaluation of medical image segmentation. J Med Imag 5(1):015006
Bártová E, Šustáčková, G, Stixová, L, Kozubek, S., Legartová, S Foltánková, V (2011) Recruitment of Oct4 protein to UV-damaged chromatin in embryonic stem cells.닏PLoS One, 6(12):e27281
Acknowledgements
Annette Schwerdtfeger for careful proofreading of the manuscript. Funding was provided by grants from the Natural Sciences and Engineering Research Council of Canada and the Neuronex Initiative (National Science Foundation, Fond de recherche du Québec – Santé). F.L.C. is a Canada Research Chair Tier II, A.B. and C.B. are both supported by a PhD scholarship from the Fonds de recherche du QuÕbec - Nature et technologies (FRQNT), an excellence scholarship from the FRQNT strategic cluster UNIQUE, and C.B. by a Leadership and Scientific Engagement Award from Université Laval.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Bilodeau, A., Bouchard, C., Lavoie-Cardinal, F. (2022). Automated Microscopy Image Segmentation and Analysis with Machine Learning. In: Heit, B. (eds) Fluorescent Microscopy. Methods in Molecular Biology, vol 2440. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2051-9_20
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2051-9_20
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2050-2
Online ISBN: 978-1-0716-2051-9
eBook Packages: Springer Protocols