Abstract
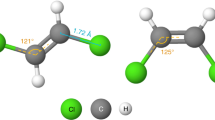
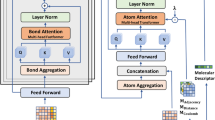
Graph Neural Networks (GNNs) play a fundamental role in many deep learning problems, in particular in cheminformatics. However, typical GNNs cannot capture the concept of chirality, which means they do not distinguish between the 3D graph of a chemical compound and its mirror image (enantiomer). The ability to distinguish between enantiomers is important especially in drug discovery because enantiomers can have very distinct biochemical properties. In this paper, we propose a theoretically justified message-passing scheme, which makes GNNs sensitive to the order of node neighbors. We apply that general concept in the context of molecular chirality to construct Chiral Edge Neural Network (ChiENN) layer which can be appended to any GNN model to enable chirality-awareness. Our experiments show that adding ChiENN layers to a GNN outperforms current state-of-the-art methods in chiral-sensitive molecular property prediction tasks.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Notes
References
Adams, K., Pattanaik, L., Coley, C.W.: Learning 3D representations of molecular chirality with invariance to bond rotations. In: The Tenth International Conference on Learning Representations, ICLR 2022, Virtual Event, 25–29 April 2022. OpenReview.net (2022)
Brown, N., Fiscato, M., Segler, M.H.S., Vaucher, A.C.: GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59(3), 1096–1108 (2019)
Chen, S., Jung, Y.: Deep retrosynthetic reaction prediction using local reactivity and global attention. JACS Au 1(10), 1612–1620 (2021)
Choukroun, Y., Wolf, L.: Geometric transformer for end-to-end molecule properties prediction. ar**v preprint ar**v:2110.13721 (2021)
Coors, B., Condurache, A.P., Geiger, A.: Spherenet: learning spherical representations for detection and classification in omnidirectional images. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 518–533 (2018)
Ganea, O., et al.: Geomol: torsional geometric generation of molecular 3D conformer ensembles. Adv. Neural. Inf. Process. Syst. 34, 13757–13769 (2021)
Gasteiger, J., Becker, F., Günnemann, S.: Gemnet: universal directional graph neural networks for molecules. Adv. Neural. Inf. Process. Syst. 34, 6790–6802 (2021)
Gasteiger, J., Groß, J., Günnemann, S.: Directional message passing for molecular graphs. ar**v preprint ar**v:2003.03123 (2020)
Gilmer, J., Schoenholz, S.S., Riley, P.F., Vinyals, O., Dahl, G.E.: Neural message passing for quantum chemistry. In: International Conference on Machine Learning, pp. 1263–1272. PMLR (2017)
Gómez-Hortigüela, L., Bernardo-Maestro, B.: Chiral organic structure-directing agents. In: Gómez-Hortigüela, L. (ed.) Insights into the Chemistry of Organic Structure-Directing Agents in the Synthesis of Zeolitic Materials. SB, vol. 175, pp. 201–244. Springer, Cham (2017). https://doi.org/10.1007/430_2017_9
Hawkins, P.C., Skillman, A.G., Warren, G.L., Ellingson, B.A., Stahl, M.T.: Conformer generation with omega: algorithm and validation using high quality structures from the protein databank and Cambridge structural database. J. Chem. Inf. Model. 50(4), 572–584 (2010)
Jamali, F., Mehvar, R., Pasutto, F.: Enantioselective aspects of drug action and disposition: therapeutic pitfalls. J. Pharm. Sci. 78, 695–715 (1989)
Kovatcheva, A., Golbraikh, A., Oloff, S., Feng, J., Zheng, W., Tropsha, A.: QSAR modeling of datasets with enantioselective compounds using chirality sensitive molecular descriptors. SAR QSAR Environ. Res. 16(1–2), 93–102 (2005)
Kreuzer, D., Beaini, D., Hamilton, W., Létourneau, V., Tossou, P.: Rethinking graph transformers with spectral attention. Adv. Neural. Inf. Process. Syst. 34, 21618–21629 (2021)
Krstulović, A.M.: Chiral Separations by HPLC. Ellis Horwood, Chichester (1989)
Liao, K., et al.: Design of catalysts for site-selective and enantioselective functionalization of non-activated primary C-H bonds. Nat. Chem. 10, 1048–1055 (2018)
Liu, C., Korablyov, M., Jastrzebski, S., Wlodarczyk-Pruszynski, P., Bengio, Y., Segler, M.H.S.: RetroGNN: fast estimation of synthesizability for virtual screening and de novo design by learning from slow retrosynthesis software. J. Chem. Inf. Model. 62(10), 2293–2300 (2022)
Mamede, R., de Almeida, B.S., Chen, M., Zhang, Q., Aires-de Sousa, J.: Machine learning classification of one-chiral-center organic molecules according to optical rotation. J. Chem. Inf. Model. 61(1), 67–75 (2020)
Mansimov, E., Mahmood, O., Kang, S., Cho, K.: Molecular geometry prediction using a deep generative graph neural network. Sci. Rep. 9(1), 20381 (2019)
Maziarka, L., Danel, T., Mucha, S., Rataj, K., Tabor, J., Jastrzebski, S.: Molecule attention transformer. CoRR abs/2002.08264 (2020)
Maziarka, Ł, Pocha, A., Kaczmarczyk, J., Rataj, K., Danel, T., Warchoł, M.: Mol-CycleGAN: a generative model for molecular optimization. J. Cheminform. 12(1), 2 (2020)
Maziarz, K., et al.: Learning to extend molecular scaffolds with structural motifs. In: The Tenth International Conference on Learning Representations, ICLR 2022, Virtual Event, 25–29 April 2022. OpenReview.net (2022). https://openreview.net/forum?id=ZTsoE8G3GG
Nguyen, L., He, H., Pham-Huy, C.: Chiral drugs: an overview. Int. J. Biomed. Sci. IJBS 2, 85–100 (2006)
Pattanaik, L., Ganea, O.E., Coley, I., Jensen, K.F., Green, W.H., Coley, C.W.: Message passing networks for molecules with tetrahedral chirality. ar**v preprint ar**v:2012.00094 (2020)
Pfaltz, A., Drury, W.: Design of chiral ligands for asymmetric catalysis: from C2-symmetric P, P- and N, N-ligands to sterically and electronically nonsymmetrical P, N-ligands. Proc. Natl. Acad. Sci. USA 101, 5723–5726 (2004)
Rampášek, L., Galkin, M., Dwivedi, V.P., Luu, A.T., Wolf, G., Beaini, D.: Recipe for a general, powerful, scalable graph transformer. Adv. Neural. Inf. Process. Syst. 35, 14501–14515 (2022)
Schneider, N., Lewis, R.A., Fechner, N., Ertl, P.: Chiral cliffs: investigating the influence of chirality on binding affinity. ChemMedChem 13(13), 1315–1324 (2018)
Schütt, K.T., Sauceda, H.E., Kindermans, P.J., Tkatchenko, A., Müller, K.R.: SchNet-a deep learning architecture for molecules and materials. J. Chem. Phys. 148(24), 241722 (2018)
Subramanian, G., Ramsundar, B., Pande, V., Denny, R.A.: Computational modeling of \(\beta \)-secretase 1 (BACE-1) inhibitors using ligand based approaches. J. Chem. Inf. Model. 56(10), 1936–1949 (2016)
Veličković, P., Cucurull, G., Casanova, A., Romero, A., Lio, P., Bengio, Y.: Graph attention networks. ar**v preprint ar**v:1710.10903 (2017)
Wang, Y., Wang, J., Cao, Z., Barati Farimani, A.: Molecular contrastive learning of representations via graph neural networks. Nat. Mach. Intell. 4(3), 279–287 (2022)
Wu, Z., et al.: MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9(2), 513–530 (2018)
Xu, M., Luo, S., Bengio, Y., Peng, J., Tang, J.: Learning neural generative dynamics for molecular conformation generation. In: 9th International Conference on Learning Representations, ICLR 2021, Virtual Event, Austria, 3–7 May 2021. OpenReview.net (2021)
Yang, K., et al.: Analyzing learned molecular representations for property prediction. J. Chem. Inf. Model. 59(8), 3370–3388 (2019)
Zaheer, M., Kottur, S., Ravanbakhsh, S., Poczos, B., Salakhutdinov, R.R., Smola, A.J.: Deep sets. In: Advances in Neural Information Processing Systems, vol. 30 (2017)
Zhu, J., et al.: Unified 2D and 3D pre-training of molecular representations. In: Zhang, A., Rangwala, H. (eds.) KDD 2022: The 28th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, Washington, DC, USA, 14–18 August 2022, pp. 2626–2636. ACM (2022)
Acknowledgements
The research of J. Tabor was supported by the Foundation for Polish Science co-financed by the European Union under the European Regional Development Fund in the POIR.04.04.00-00-14DE/18-00 project carried out within the Team-Net program. The research of P. Gaiński and M. Śmieja was supported by the National Science Centre (Poland), grant no. 2022/45/B/ST6/01117.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Ethical Statement
As we consider our work to be fundamental research, there are no direct ethical risks or societal consequences; these have to be analyzed per concrete applications.
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Gaiński, P., Koziarski, M., Tabor, J., Śmieja, M. (2023). ChiENN: Embracing Molecular Chirality with Graph Neural Networks. In: Koutra, D., Plant, C., Gomez Rodriguez, M., Baralis, E., Bonchi, F. (eds) Machine Learning and Knowledge Discovery in Databases: Research Track. ECML PKDD 2023. Lecture Notes in Computer Science(), vol 14171. Springer, Cham. https://doi.org/10.1007/978-3-031-43418-1_3
Download citation
DOI: https://doi.org/10.1007/978-3-031-43418-1_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-43417-4
Online ISBN: 978-3-031-43418-1
eBook Packages: Computer ScienceComputer Science (R0)