Abstract
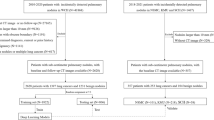
Malignant Pleural Mesothelioma (MPM) is a rare cancer associated with exposure to asbestos fibres. It grows in the pleural space surrounding the lungs, exhibiting an irregular shape with high surface-to-volume ratio. Reliable measurements are important to assessing treatment efficacy, however these tumour characteristics make manual measurements time consuming, and prone to intra- and inter-observer variation. Previously we described a fully automatic Convolutional Neural Network (CNN) for volumetric measurement of MPM in CT images, trained and evaluated by seven-fold cross validation on 123 CT datasets with expert manual annotations. The mean difference between the manual and automatic volume measurements was not significantly different from zero (27.2 cm\(^3\); \(p = 0.225\)), the 95% limits of agreement were between −417 and +363 cm\(^3\), and the mean Dice coefficient was 0.64. Previous studies have focused on images with known MPM, sometimes even focusing on the lung with known MPM. In this paper, we investigate the false positive detection rate in a large image set with no known MPM. For this, a cohort of 14,965 subjects from the National Lung Screening Trial (NLST) were analysed. The mean volume of “MPM” found in these images by the automated detector was 3.6 cm\(^3\) (compared with 547.2 cm\(^3\) for MPM positive subjects). A qualitative examination of the one hundred subjects with the largest probable false detection volumes found that none of them were normal: the majority contain hyperdense pathology, large regions of pleural effusion, or evidence of pleural thickening. One false positive was caused by liver masses. The next step will be to evaluate the automated measurement accuracy on an independent, unseen, multi-centre data set.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Abdel-Rahman, O.: Global trends in mortality from malignant mesothelioma: analysis of WHO mortality database (1994–2013). Clin. Respir. J. (2018). https://doi.org/10.1111/crj.12778
Anderson, O., et al.: Fully automated volumetric measurement of malignant pleural mesothelioma from computed tomography images by deep learning: preliminary results of an internal validation. In: BIOIMAGING 2020–7th International Conference on Bioimaging, Proceedings; Part of 13th International Joint Conference on Biomedical Engineering Systems and Technologies, BIOSTEC 2020 (2020). https://doi.org/10.5220/0008976100640073
Armato, S.G., Nowak, A.K., Francis, R.J., Kocherginsky, M., Byrne, M.J.: Observer variability in mesothelioma tumor thickness measurements: defining minimally measurable lesions. J. Thoracic Oncol. (2014). https://doi.org/10.1097/JTO.0000000000000211
Attanoos, R.L., Gibbs, A.R.: Pathology of malignant mesothelioma. Histopathology 30(5), 403–418 (1997). https://doi.org/10.1046/j.1365-2559.1997.5460776.x. https://onlinelibrary.wiley.com/doi/abs/10.1046/j.1365-2559.1997.5460776.x
Bianchi, C., Giarelli, L., Grandi, G., Brollo, A., Ramani, L., Zuch, C.: Latency periods in asbestos-related mesothelioma of the pleura. Eur. J. Cancer Prev. 6, 162–166 (1997)
Blyth, K., et al.: An update regarding the Prediction of ResIstance to chemotherapy using Somatic copy number variation in Mesothelioma (PRISM) study. Lung Cancer (2018). https://doi.org/10.1016/s0169-5002(18)30090-4
Brahim, W., Mestiri, M., Betrouni, N., Hamrouni, K.: Malignant pleural mesothelioma segmentation for photodynamic therapy planning. Comput. Med. Imaging Graph. (2018). https://doi.org/10.1016/j.compmedimag.2017.05.006
Byrne, M.J., Nowak, A.K.: Modified RECIST criteria for assessment of response in malignant pleural mesothelioma. Ann. Oncol. (2004). https://doi.org/10.1093/annonc/mdh059
Chen, M., Helm, E., Joshi, N., Gleeson, F., Brady, M.: Computer-aided volumetric assessment of malignant pleural mesothelioma on CT using a random walk-based method. Int. J. Comput. Assist. Radiol. Surg. 12(4), 529–538 (2016). https://doi.org/10.1007/s11548-016-1511-3
Chollet, F.: Keras (2015). https://keras.io/
Eisenhauer, E.A., et al.: New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur. J. Cancer (2009). https://doi.org/10.1016/j.ejca.2008.10.026
Gudmundsson, E., Straus, C., Li, F., Kindler, H., Armato, S.: P1.06-04 deep learning-based segmentation of mesothelioma on CT scans: application to patient scans exhibiting pleural effusion. J. Thoracic Oncol. (2019). https://doi.org/10.1016/j.jtho.2019.08.991
Gudmundsson, E., Straus, C.M., Armato, S.G.: Deep convolutional neural networks for the automated segmentation of malignant pleural mesothelioma on computed tomography scans. J. Med. Imaging (2018). https://doi.org/10.1117/1.jmi.5.3.034503
Gudmundsson, E., Straus, C.M., Li, F., Armato, S.G.: Deep learning-based segmentation of malignant pleural mesothelioma tumor on computed tomography scans: application to scans demonstrating pleural effusion. J. Med. Imaging (2020). https://doi.org/10.1117/1.jmi.7.1.012705
Ioffe, S., Szegedy, C.: Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the International Conference on Machine Learning (ICML). Proceedings of Machine Learning Research, vol. 37, pp. 448–456 (2015). http://proceedings.mlr.press/v37/ioffe15.html
Deng, J., Dong, W., Socher, R., Li, L.-J., Li, K., Fei-Fei, L.: ImageNet: a large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition (2009). https://doi.org/10.1109/CVPRW.2009.5206848
Labby, Z.E., et al.: Variability of tumor area measurements for response assessment in malignant pleural mesothelioma. Med. Phys. (2013). https://doi.org/10.1118/1.4810940
Martin Bland, J., Altman, D.G.: Statistical methods for assessing agreement between two methods of clinical measurement. The Lancet (1986). https://doi.org/10.1016/S0140-6736(86)90837-8
National Lung Screening Trial Research Team: The national lung screening trial: overview and study design. Radiology (2011). https://doi.org/10.1148/radiol.10091808
Ng, C.S., Munden, R.F., Libshitz, H.I.: Malignant pleural mesothelioma: the spectrum of manifestations on CT in 70 cases. Clin. Radiol. (1999). https://doi.org/10.1016/S0009-9260(99)90824-3
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (2015). https://doi.org/10.1007/978-3-319-24574-4_28. http://arxiv.org/abs/1505.04597
Schwartz, L.H., et al.: RECIST 1.1 - Update and clarification: from the RECIST committee. Eur. J. Cancer (2016). https://doi.org/10.1016/j.ejca.2016.03.081
Sensakovic, W.F., et al.: Computerized segmentation and measurement of malignant pleural mesothelioma. Med. Phys. (2011). https://doi.org/10.1118/1.3525836
Smith, L.N.: Cyclical learning rates for training neural networks. In: IEEE Winter Conference on Applications of Computer Vision (2017). https://doi.org/10.1109/WACV.2017.58
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I., Salakhutdinov, R.: Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014). https://doi.org/10.1214/12-AOS1000
Tsim, S., et al.: Diagnostic and Prognostic Biomarkers in the Rational Assessment of Mesothelioma (DIAPHRAGM) study: Protocol of a prospective, multicentre, observational study. BMJ Open (2016). https://doi.org/10.1136/bmjopen-2016-013324
Yoon, S.H., Kim, K.W., Goo, J.M., Kim, D.W., Hahn, S.: Observer variability in RECIST-based tumour burden measurements: a meta-analysis. Eur. J. Cancer 53, 5–15 (2016). https://doi.org/10.1016/j.ejca.2015.10.014
Acknowledgments
The authors thank the National Cancer Institute for access to NCI’s data collected by the National Lung Screening Trial (NLST). The statements contained herein are solely those of the authors and do not represent or imply concurrence or endorsement by NCI.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Anderson, O. et al. (2021). Estimating the False Positive Prediction Rate in Automated Volumetric Measurements of Malignant Pleural Mesothelioma. In: Ye, X., et al. Biomedical Engineering Systems and Technologies. BIOSTEC 2020. Communications in Computer and Information Science, vol 1400. Springer, Cham. https://doi.org/10.1007/978-3-030-72379-8_7
Download citation
DOI: https://doi.org/10.1007/978-3-030-72379-8_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-72378-1
Online ISBN: 978-3-030-72379-8
eBook Packages: Computer ScienceComputer Science (R0)