Abstract
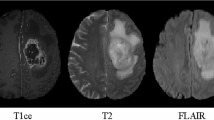
Segmentation of brain tumors and their subregions remains a challenging task due to their weak features and deformable shapes. In this paper, three patterns (cross-skip, skip-1 and skip-2) of distributed dense connections (DDCs) are proposed to enhance feature reuse and propagation of CNNs by constructing tunnels between key layers of the network. For better detecting and segmenting brain tumors from multi-modal 3D MR images, CNN-based models embedded with DDCs (DDU-Nets) are trained efficiently from pixel to pixel with a limited number of parameters. Postprocessing is then applied to refine the segmentation results by reducing the false-positive samples. The proposed method is evaluated on the BraTS 2019 dataset with results demonstrating the effectiveness of the DDU-Nets while requiring less computational cost.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017)
Bakas, S., et al.: Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive 286 (2017)
Bakas, S., et al.: Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. In: Scientific Data (2017)
Bakas, S., et al.: Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. ar**v preprint ar**v:1811.02629 (2018)
Cao, H., Bernard, S., Heutte, L., Sabourin, R.: Improve the performance of transfer learning without fine-tuning using dissimilarity-based multi-view learning for breast cancer histology images. In: Campilho, A., Karray, F., ter Haar Romeny, B. (eds.) ICIAR 2018. LNCS, vol. 10882, pp. 779–787. Springer, Cham (2018). https://doi.org/10.1007/978-3-319-93000-8_88
Dong, H., Yang, G., Liu, F., Mo, Y., Guo, Y.: Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In: Valdés Hernández, M., González-Castro, V. (eds.) MIUA 2017. CCIS, vol. 723, pp. 506–517. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-60964-5_44
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., Maier-Hein, K.H.: No new-net. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 234–244. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_21
Larsson, G., Maire, M., Shakhnarovich, G.: FractalNet: ultra-deep neural networks without residuals. ar**v preprint ar**v:1605.07648 (2016)
Li, X., Chen, H., Qi, X., Dou, Q., Fu, C.W., Heng, P.A.: H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med. Imaging 37(12), 2663–2674 (2018)
Mamelak, A.N., Jacoby, D.B.: Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601). Expert Opin. Drug Deliv. 4(2), 175–186 (2007)
Menze, B.H., et al.: The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans. Med. Imaging 34(10), 1993–2024 (2014)
Myronenko, A.: 3D MRI brain tumor segmentation using autoencoder regularization. In: Crimi, A., Bakas, S., Kuijf, H., Keyvan, F., Reyes, M., van Walsum, T. (eds.) BrainLes 2018. LNCS, vol. 11384, pp. 311–320. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-11726-9_28
Pleiss, G., Chen, D., Huang, G., Li, T., van der Maaten, L., Weinberger, K.Q.: Memory-efficient implementation of DenseNets. ar**v preprint ar**v:1707.06990 (2017)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Zhang, H., Li, J., Shen, M., Wang, Y., Yang, GZ. (2020). DDU-Nets: Distributed Dense Model for 3D MRI Brain Tumor Segmentation. In: Crimi, A., Bakas, S. (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2019. Lecture Notes in Computer Science(), vol 11993. Springer, Cham. https://doi.org/10.1007/978-3-030-46643-5_20
Download citation
DOI: https://doi.org/10.1007/978-3-030-46643-5_20
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-46642-8
Online ISBN: 978-3-030-46643-5
eBook Packages: Computer ScienceComputer Science (R0)