Abstract
Background
In the era of molecular stratification and effective multimodality therapies, surgical staging of the axilla is becoming less relevant for patients with estrogen receptor (ER)-positive early-stage breast cancer (EBC). Therefore, a nonsurgical method for accurately predicting lymph node disease is the next step in the de-escalation of axillary surgery. This study sought to identify epigenetic signatures in the primary tumor that accurately predict lymph node status.
Patients and Methods
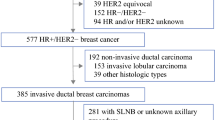
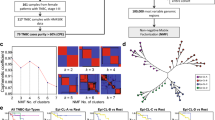
We selected a cohort of patients in The Cancer Genome Atlas (TCGA) with ER-positive, HER2-negative invasive ductal carcinomas, and clinically-negative axillae (n = 127). Clinicopathological nomograms from the Memorial Sloan Kettering Cancer Center (MSKCC) and the MD Anderson Cancer Center (MDACC) were calculated. DNA methylation (DNAm) patterns from primary tumor specimens were compared between patients with pN0 and those with > pN0. The cohort was divided into training (n = 85) and validation (n = 42) sets. Random forest was employed to obtain the combinations of DNAm features with the highest accuracy for stratifying patients with > pN0. The most efficient combinations were selected according to the area under the curve (AUC).
Results
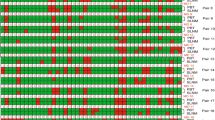
Clinicopathological models displayed a modest predictive potential for identifying > pN0 disease (MSKCC AUC 0.76, MDACC AUC 0.69, p = 0.15). Differentially methylated sites (DMS) between patients with pN0 and those with > pN0 were identified (n = 1656). DMS showed a similar performance to the MSKCC model (AUC = 0.76, p = 0.83). Machine learning approaches generated five epigenetic classifiers, which showed higher discriminative potential than the clinicopathological variables tested (AUC > 0.88, p < 0.05).
Conclusions
Epigenetic classifiers based on primary tumor characteristics can efficiently stratify patients with no lymph node involvement from those with axillary lymph node disease, thereby providing an accurate method of staging the axilla.



Similar content being viewed by others
References
Society of Surgical Oncology. Choosing Wisely: Don’t routinely use sentinel node biopsy in clinically node negative women ≥ 70 years of age with early stage hormone receptor positive, HER2 negative invasive breast cancer. https://www.choosingwisely.org/clinician-lists/sso-sentinel-node-biopsy-in-node-negative-women-70-and-over/. Updated 07/27/2021. Accessed 8 March 2022.
National Comprehensive Cancer Network. Breast Cancer (Version 3.2022). https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Accessed 18 May 2022.
Giuliano AE, Ballman KV, McCall L, et al. Effect of axillary dissection vs no axillary dissection on 10-year overall survival among women with invasive breast cancer and sentinel node metastasis: the ACOSOG Z0011 (Alliance) randomized clinical trial. JAMA. 2017;318(10):918–26.
Kalinsky K, Barlow WE, Gralow JR, et al. 21-gene assay to inform chemotherapy benefit in node-positive breast cancer. N Engl J Med. 2021;385(25):2336–47.
Veronesi U, Paganelli G, Viale G, et al. A randomized comparison of sentinel-node biopsy with routine axillary dissection in breast cancer. N Engl J Med. 2003;349(6):546–53.
Krag DN, Anderson SJ, Julian TB, et al. Sentinel-lymph-node resection compared with conventional axillary-lymph-node dissection in clinically node-negative patients with breast cancer: overall survival findings from the NSABP B-32 randomised phase 3 trial. Lancet Oncol. 2010;11(10):927–33.
Bevilacqua JL, Kattan MW, Fey JV, Cody HS 3rd, Borgen PI, Van Zee KJ. Doctor, what are my chances of having a positive sentinel node? A validated nomogram for risk estimation. J Clin Oncol. 2007;25(24):3670–9.
Klar M, Foeldi M, Markert S, Gitsch G, Stickeler E, Watermann D. Good prediction of the likelihood for sentinel lymph node metastasis by using the MSKCC nomogram in a German breast cancer population. Ann Surg Oncol. 2009;16(5):1136–42.
Chen JY, Chen JJ, Yang BL, et al. Predicting sentinel lymph node metastasis in a Chinese breast cancer population: assessment of an existing nomogram and a new predictive nomogram. Breast Cancer Res Treat. 2012;135(3):839–48.
Reyal F, Rouzier R, Depont-Hazelzet B, et al. The molecular subtype classification is a determinant of sentinel node positivity in early breast carcinoma. PLoS One. 2011;6(5):e20297.
Orozco JIJ, Knijnenburg TA, Manughian-Peter AO, et al. Epigenetic profiling for the molecular classification of metastatic brain tumors. Nat Commun. 2018;9(1):4627.
DiNome ML, Orozco JIJ, Matsuba C, et al. Clinicopathological features of triple-negative breast cancer epigenetic subtypes. Ann Surg Oncol. 2019;26(10):3344–53.
Urrutia G, Laurito S, Marzese DM, et al. Epigenetic variations in breast cancer progression to lymph node metastasis. Clin Exp Metastasis. 2015;32(2):99–110.
Salomon MP, Orozco JIJ, Wilmott JS, et al. Brain metastasis DNA methylomes, a novel resource for the identification of biological and clinical features. Sci Data. 2018;5:180245.
National Cancer Institute/GDC Data Portal. Harmonized Cancer Datasets. Genomic Data Commons Data Portal. https://portal.gdc.cancer.gov. Accessed 1 August 2020.
Memorial Sloan Kettering Cancer Center. Breast cancer nomogram: sentinel lymph node metastasis. http://nomograms.mskcc.org/breast/BreastSLNodeMetastasisPage.aspx. Accessed 9 November 2020.
MD Anderson Cancer Center. Breast cancer nomogram to predict positive sentinel lymph nodes, without neoadjuvant chemotherapy. http://www3.mdanderson.org/app/medcalc/bc_nomogram3/index.cfm?pagename=sln. Accessed 9 November 2020.
Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71.
Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun. 2015;6:8971.
John CR, Watson D, Russ D, et al. M3C: monte carlo reference-based consensus clustering. Sci Rep. 2020;10(1):1816.
O’Connor T, Grant CE, Bodén M, Bailey TL. T-gene: improved target gene prediction. Bioinformatics. 2020;36(12):3902–4.
Grote S. GOfuncR: Gene ontology enrichment using FUNC. R package version 1.16.0. 2022.
Diaz-Uriarte R. GeneSrF and varSelRF: a web-based tool and R package for gene selection and classification using random forest. BMC Bioinform. 2007;8:328.
Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21(20):3940–1.
DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45.
DeSantis CE, Ma J, Gaudet MM, et al. Breast cancer statistics. CA Cancer J Clin. 2019;69(6):438–51.
Donker M, van Tienhoven G, Straver ME, et al. Radiotherapy or surgery of the axilla after a positive sentinel node in breast cancer (EORTC 10981–22023 AMAROS): a randomised, multicentre, open-label, phase 3 non-inferiority trial. Lancet Oncol. 2014;15(12):1303–10.
Qiu PF, Liu JJ, Wang YS, et al. Risk factors for sentinel lymph node metastasis and validation study of the MSKCC nomogram in breast cancer patients. Jpn J Clin Oncol. 2012;42(11):1002–7.
Moran S, Martínez-Cardús A, Sayols S, et al. Epigenetic profiling to classify cancer of unknown primary: a multicentre, retrospective analysis. Lancet Oncol. 2016;17(10):1386–95.
Capper D, Jones DTW, Sill M, et al. DNA methylation-based classification of central nervous system tumours. Nature. 2018;555(7697):469–74.
Klughammer J, Kiesel B, Roetzer T, et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat Med. 2018;24(10):1611–24.
Jeschke J, Bizet M, Desmedt C, et al. DNA methylation-based immune response signature improves patient diagnosis in multiple cancers. J Clin Invest. 2017;127(8):3090–102.
Ensenyat-Mendez M, Íñiguez-Muñoz S, Sesé B, Marzese DM. iGlioSub: an integrative transcriptomic and epigenomic classifier for glioblastoma molecular subtypes. BioData Min. 2021;14(1):42.
McKinney SM, Sieniek M, Godbole V, et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577(7788):89–94.
Møller M, Strand SH, Mundbjerg K, et al. Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Sci Rep. 2017;7:40636.
Uhl B, Gevensleben H, Tolkach Y, et al. PITX2 DNA methylation as biomarker for individualized risk assessment of prostate cancer in core biopsies. J Mol Diagn. 2017;19(1):107–14.
Acknowledgment
The research described was supported by NIH/National Center for Advancing Translational Science (NCATS) UCLA CTSI Grant No. UL1TR001881, the Associates for Breast and Prostate Cancer Studies (ABCs) Foundation, the Fashion Footwear Association of New York (FFANY) Foundation, the Spanish Instituto de la Salud Carlos III (#CP17/0018), co-funded by ERDF “A way to make Europe,” the Asociación Española Contra el Cancer (AECC), and the UCLA Breast Cancer Epigenetics Research Program.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
The authors have no conflict of interest disclosures to report.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Orozco, J.I.J., Le, J., Ensenyat-Mendez, M. et al. Machine Learning-Based Epigenetic Classifiers for Axillary Staging of Patients with ER-Positive Early-Stage Breast Cancer. Ann Surg Oncol 29, 6407–6414 (2022). https://doi.org/10.1245/s10434-022-12143-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-12143-6