Abstract
Background
To identify potential serum biomarkers for differentiating between axial psoriatic arthritis (axPsA) and peripheral psoriatic arthritis (pPsA).
Methods
Serum samples were collected from patients with PsA to create a biomarker discovery cohort and a verification cohort. Patients with PsA were classified into axial or peripheral subtypes based on imaging criteria. Untargeted proteomics technology was used in the discovery phase to screen for biomarkers, and candidate biomarkers were evaluated using enzyme-linked immunosorbent assay (ELISA) in the verification phase.
Results
We identified 45 significantly differentially expressed proteins (DEPs) between axPsA (n = 20) and pPsA (n = 20) with liquid chromatography-mass spectrometry. Among these DEPs, serum pigment epithelium-derived factor (PEDF) was identified as a candidate biomarker using the Boruta algorithm and lasso regression. Results of ELISA further confirmed that the level of serum PEDF expression was significantly higher in axPsA (n = 37) than in pPsA (n = 51) at the verification cohort (37.9 ± 10.1 vs. 30.5 ± 8.9 μg/mL, p < 0.001). Receiver operating characteristics analysis showed that PEDF had an area under the curve (AUC) of 0.72. Serum PEDF was positively correlated with body mass index and C-reactive protein. Additionally, there was a tendency towards a positive correlation between PEDF and the Bath Ankylosing Spondylitis Disease Activity Index.
Conclusions
This study provided a comprehensive characterization of the proteome in axPsA and pPsA and identified a candidate biomarker, PEDF, that may contribute to early diagnosis for axPsA.
Similar content being viewed by others
Background
Psoriatic arthritis (PsA) is a musculoskeletal disease that can affect multiple domains, accompanied by other comorbidities such as metabolic syndrome. Some patients with PsA may exhibit axial involvement, referred to as axial PsA (axPsA), while those without axial joint involvement are known as peripheral PsA (pPsA). There is an ongoing controversy regarding whether axial PsA is a distinct disease from axial Spondyloarthritis (axSpA), but multiple studies have pointed out clinical distinctions between axPsA and axSpA [1]. These differences include a less common presence of Inflammatory back pain (IBP), lower rates of HLA-B27 positivity, characteristic axial imaging changes, and so on [2]. IBP, a critical symptom of inflammation in the axial joints for axSpA diagnosis, is found in only 26% of axPsA patients who meet imaging criteria [3]. Secondly, unlike the high positivity rate of HLA-B27 in ankylosing spondylitis [4], the rate of HLA-B27 positivity in patients with PsA is only 20–35% [5, 6], whereas in axPsA this number rises to 43% [7]. All these differences from axSpA may delay the identification of axial involvement in PsA. Conventional synthetic disease-modifying antirheumatic drugs (csDMARDs), which are commonly used in the treatment of pPsA, are considered ineffective for treating axial disease. Guidelines recommend that in patients with predominantly axial disease which is active and has insufficient response to nonsteroidal anti-inflammatory drugs (NSAIDs), therapy with biological disease-modifying antirheumatic drugs (bDMARDs) should be considered [8]. Misdiagnosis of axPsA can result in the progression of axial involvement, leading to joint damage and a significant impact on prognosis [9]. Besides, some patients with PsA only develop axial disease in the late stage of the disease [10, 11], which also implies the need to pay attention to screening during the follow-up period. However, the frequent use of imaging exams undoubtedly increases the radiation and economic burden on patients.
The utilization of reliable biomarkers aids in the early diagnosis of diseases and understanding of the pathogenesis of diseases [12, 13]. Therefore, the discovery of a reliable biomarker in patients with PsA for predicting or identifying involvement of the axial disease would greatly benefit clinicians in prescribing imaging examinations and initiating early use of biologic agents. This, in turn, would improve the prognosis, physical function, and overall quality of life for these patients. Multiple types of samples, such as urine, feces, and saliva can be used as potential samples for research; however, serum and plasma are commonly used due to their ease of collection and stable composition [14]. Techniques for analyzing the serum proteome include mass spectrometry (MS), Multiplex bead- or aptamer-based assays (Slow off-rate modified aptamer scan), and Proximity extension assay (Olink) [15]. Although the latter two techniques offer higher sensitivity, as targeted proteomic techniques, they only monitor the presence or absence of target proteins, whereas MS techniques allow for the hypothesis-free approach with shotgun untargeted MS workflows [16]. Therefore, this study employed untargeted proteomic technology to explore serum biomarkers of axPsA.
Methods
Study design and collection of clinical samples
The research study was divided into two phases: the discovery phase and the verification phase. The workflow of the study is illustrated in Fig. 1. The study included patients who were diagnosed with PsA based on the Classification Criteria for Psoriatic Arthritis (CASPAR) [17] and also had accessible results of sacroiliac joint computerized tomography (CT) and spinal X-ray, for the formation of the discovery and verification cohorts. The definition of axPsA was based on a previous study [11], which included the presence of New York criteria sacroiliitis (unilateral grade ≥ 3, or bilateral grade ≥ 2 sacroiliitis), and/or≥ 1 marginal/paramarginal syndesmophytes of the cervical or lumbar spine. The grading of sacroiliac arthritis on CT examination was done according to the study by Ye et al. [2b).
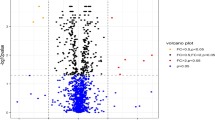
Unbiased L-MS/MS-based protein analysis. a PCA score plot of the serum samples of the discovery cohort. b PLS-DA score plot of the serum samples of the discovery cohort. c This Venn diagram shows the number of DEPs found in the pairwise comparison among the three groups. d The volcano plot shows the DEPs between axPsA and pPsA. PEDF was the top-upregulated DEP according to the p-value. axPsA axial psoriatic arthritis, pPsA peripheral psoriatic arthritis, HC healthy control, DEPs differentially expressed proteins, PEDF pigment epithelium-derived factor
We identified a total of 130 DEPs when comparing healthy controls and all patients with PsA. When comparing axPsA and HC, pPsA and HC, and axPsA and pPsA, we identified 101, 120, and 45 DEPs, respectively. The overlap between these DEP group sets was analyzed with the Venn diagram, which is illustrated in Fig. 2c. Among these 45 DEPs between axPsA and pPsA, 11 proteins were found to be up-regulated, while 34 proteins were down-regulated in axPsA. A volcano plot was used to visualize the changes in protein expression between axPsA and pPsA, as shown in Fig. 2d. Among these proteins, PEDF was found to be the top significantly upregulated protein in axPsA, based on the adjusted P value.
Using the bioinformatics method, we conducted an analysis of the DEPs between axPsA and pPsA. The results of the GO analysis indicated that the DEPs were associated with biological processes related to innate immunity like complement, coagulation, and the regulation of proteolytic activity. Furthermore, these DEPs were involved in the regulation of enzyme activity, which was identified as the most important molecular function. KEGG analysis showed that these DEPs were mainly involved in the complement pathway and hemostasis (Additional file 1: Figure S1).
Identification of candidate biomarkers
To better identify clinically available biomarkers, we employed various approaches to select candidate biomarkers. Boruta analyses confirmed 28 biomarkers that are important for identifying patients. Random Forest (RF) analyses were conducted to objectively evaluate the importance of serum proteins, and the top ten proteins are displayed in Fig.3b, c. Additionally, LASSO regression selected 7 DEPs. Among these candidate biomarkers, PEDF appeared in all lists and ranked first in RF. Therefore, we selected PEDF as a potential marker for further verification. Based on quantitative analysis using mass spectrometry data, we observed a significant upregulation of PEDF in axPsA compared to pPsA. Receiver operating characteristic (ROC) analyses of PEDF yielded an area under the curve (AUC) value of 0.925.
Identification of candidate biomarkers based on MS data from the discovery cohort. a Feature selection based on the Buruta algorithm; Feature ordering based on mean decrease accuracy (b) and mean decrease gini (c) in random forest model. PEDF was the top rank DEP; d quantitative analysis of serum PEDF levels in two groups using mass spectrometry data (****p < 0.001). The intensity of PEDF was normalized. e Receiver operating characteristic curve analysis of candidate biomarkers for axPsA vs. pPsA based on quantitative analysis of mass spectrometry data. PEDF pigment epithelium-derived factor, DEPs differentially expressed proteins, axPsA axial psoriatic arthritis, pPsA peripheral psoriatic arthritis
ELISA verification of MS-identified biomarkers
To further investigate the expression of PEDF in the serum of patients with axPsA and pPsA, we detected the level of PEDF in 37 patients with axPsA, and 51 patients with pPsA by ELISA in the verification phase. As shown in Fig. 4, PEDF expression was significantly higher in axPsA compared with pPsA (37.9 ± 10.1 vs. 30.5 ± 8.9 μg/mL, p < 0.001), the AUC score was 0.72 (95%CI 0.61–0.83). PEDF, BMI, and the clinical variables with significant differences in univariate analysis were included in the multivariate analysis. The results showed that PEDF remained significantly elevated in axPsA patients (P = 0.017, Additional file 1: Table S1). There were no significant differences observed in the level of PEDF among axPsA patients with different imaging types (Additional file 1: Figure S2). We performed a correlation analysis between the serum levels of PEDF and major disease manifestations in patients with PsA. The results showed that serum PEDF was positively correlated with BMI (r = 0.4, P < 0.001) and CRP (r = 0.42, P < 0.001). Additionally, a noticeable trend towards a positive correlation between PEDF and Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) was observed (r = 0.36, P = 0.064). There were no significant correlations found between serum PEDF levels and swollen joint count (SJC), TJC, and psoriasis area and severity index (PASI) (Fig. 5).
The level of PEDF in the patients in the verification cohort. a The comparison of serum PEDF between axPsA and pPsA with ELISA. (***p < 0.001). b Receiver operating characteristic curve analysis of candidate biomarkers for axPsA vs. pPsA. PEDF pigment epithelium-derived factor, axPsA axial psoriatic arthritis, pPsA peripheral psoriatic arthritis
Correlation analysis between the serum level of PEDF and major disease manifestations. The correlation between BMI (a), CRP (b), ESR (c), SJC (d), TJC (e), PASI (f), BASFI (g), BASDAI (h), and serum PEDF. The data of BASDAI and BASFI was only available in patients with axPsA. BMI body mass index, CRP C reactive protein ESR erythrocyte sedimentation rate, SJC swollen joints count, TJC tender joints count, PASI Psoriasis Area and Severity Index, BASFI Bath AS Functional Index, BASDAI Bath Ankylosing Spondylitis Disease Activity Index
Discussion
In recent years, there has been a growing interest in studying axPsA. Several studies have identified distinct characteristics that differentiate axPsA from axSpA, which may delay diagnosis and treatment [20, 21]. While reliable biomarkers can aid clinicians in prescribing more targeted imaging tests and identifying asymptomatic individuals with axial involvement, it is imperative to ensure that these patients receive timely diagnoses and appropriate treatments, such as interleukin-17A inhibitors. These inhibitors have been proven to delay radiographic progression and prevent loss of function [22]. Thus, this study utilized mass spectrometry technology to explore biomarkers capable of distinguishing between axPsA and pPsA and confirmed the dependability of serum PEDF as a potential biomarker.
As we all know, the efficacy of a biomarker is related to the definition of the disease. An important initiative in this field is the Axial Involvement in Psoriatic Arthritis cohort (AXIS) study, which aims to establish classification criteria for axPsA [23]. Considering the objectivity of imaging examinations and the poor sensitivity and specificity of existing IBP criteria in patients with axPsA [24], we refer to the imaging criteria used in previous studies. For the identification of axPsA in our study, we employed imaging criteria from previous research, including the New York criteria for sacroiliitis and/or syndesmophyte of the spine [11]. Given the high accessibility of sacroiliac joint CT in our cohort, a large number of patients had access to this imaging data. Additionally, several studies have shown that CT demonstrates superior diagnostic accuracy for axSpA [25]. Another study comparing magnetic resonance imaging and CT evaluations of axial lesions in the sacroiliac joint also indicated that CT has excellent specificity and good sensitivity [39]. A suite of machine learning techniques, such as logistic regression, random forests, and support vector machines can be used in the identification of a multivariate biomarker panel [40]. Thirdly, the patients included in this study were not treatment-naive, and the therapeutic medications may have influenced the outcomes. However, the unselected patients in this study align more closely with the real clinical environment and are more conducive to clinical applicability.
Conclusions
In conclusion, we utilized mass spectrometry to analyze the serum proteome in patients with axPsA and pPsA, and identified several DEPs between the two groups. AxPsA and pPsA have distinct serum protein profiles that can be used as biomarkers to discriminate between them. Among these proteins, PEDF showed promise as a potential biomarker, and its validity was confirmed using ELISA in a larger verification cohort. However, further validation is still needed in patients from an expanded or independent cohort before it becomes a truly reliable marker for clinical practice. Additionally, considering the clinical heterogeneity and potential comorbidities in patients with PsA, a biomarker panel with multiple proteins may be a more ideal diagnostic tool.
Availability of data and materials
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD044598 (Accession Number: IPX0006903001).
Abbreviations
- AUC:
-
Area under the curve
- axPsA:
-
Axial psoriatic arthritis
- axSpA:
-
Axial spondyloarthritis
- BASDAI:
-
Bath ankylosing spondylitis disease activity index
- BASFI:
-
Bath ankylosing spondylitis functional index
- bDMARDs:
-
Biological disease-modifying antirheumatic drugs
- BMI:
-
Body mass index
- CASPAR:
-
Classification criteria for psoriatic arthritis
- CRP:
-
C-reactive protein
- csDMRADs:
-
Conventional synthetic disease-modifying antirheumatic drugs
- CT:
-
Computerized tomography
- DAPSA:
-
Disease activity index for psoriatic arthritis
- DAS28:
-
Disease activity score 28
- DDA:
-
Data-dependent acquisition
- DEPs:
-
Differentially expressed proteins
- DIA:
-
Data-independent acquisition
- ELISA:
-
Enzyme-linked immunosorbent assay
- ESR:
-
Erythrocyte sedimentation rate
- FA:
-
Formic acid
- GO:
-
Gene ontology
- HC:
-
Healthy control
- LASSO:
-
Least absolute shrinkage and selection operator
- IBD:
-
Inflammatory bowel disease
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- MS:
-
Mass spectrometry
- NSAIDs:
-
Nonsteroidal anti-inflammatory drugs
- PASI:
-
Psoriasis area and severity index
- PCA:
-
Principal component analysis
- PEDF:
-
Pigment epithelium-derived factor
- PLS-DA:
-
Pleast squares-discriminant analysis
- pPsA:
-
Peripheral psoriatic arthritis
- PsA:
-
Psoriatic arthritis
- ROC:
-
Operating characteristic
- SJC:
-
Swollen joint count
- TJC:
-
Tender joint count
- VEGF:
-
Vascular endothelial growth factor
References
Feld J, Chandran V, Haroon N, Inman R, Gladman D. Axial disease in psoriatic arthritis and ankylosing spondylitis: a critical comparison. Nat Rev Rheumatol. 2018;14(6):363–71.
Gladman DD. Axial psoriatic arthritis. Curr Rheumatol Rep. 2021;23(6):35.
Elsawy NA, Farahat SE, Shafei MME, Elnemr R. Axial involvement in psoriatic arthritis: a cross-sectional clinical and radiologic studies. Curr Rheumatol Rev. 2023. https://doi.org/10.2174/1573397119666230911111023.
Brown MA, Kenna T, Wordsworth BP. Genetics of ankylosing spondylitis—insights into pathogenesis. Nat Rev Rheumatol. 2016;12(2):81–91.
Queiro R, Torre JC, Gonzalez S, Lopez-Larrea C, Tinture T, Lopez-Lagunas I. HLA antigens may influence the age of onset of psoriasis and psoriatic arthritis. J Rheumatol. 2003;30(3):505–7.
Haroon M, Winchester R, Giles JT, Heffernan E, FitzGerald O. Certain class I HLA alleles and haplotypes implicated in susceptibility play a role in determining specific features of the psoriatic arthritis phenotype. Ann Rheum Dis. 2016;75(1):155–62.
Marsal S, Armadans-Gil L, Martinez M, Gallardo D, Ribera A, Lience E. Clinical, radiographic and HLA associations as markers for different patterns of psoriatic arthritis. Rheumatology. 1999;38(4):332–7.
Gossec L, Baraliakos X, Kerschbaumer A, de Wit M, McInnes I, Dougados M, et al. EULAR recommendations for the management of psoriatic arthritis with pharmacological therapies: 2019 update. Ann Rheum Dis. 2020;79(6):700–12.
Santos H, Henriques AR, Machado PM, Lopez-Medina C, Dougados M, Canhão H, et al. Determinants of health-related quality of life and global functioning and health in axSpA, pSpA, and PsA: results from the ASAS-PerSpA study. Rheumatology. 2023. https://doi.org/10.1093/rheumatology/kead503.
Queiro R, Belzunegui J, González C, De DJ, Sarasqueta C, Torre JC, et al. Clinically asymptomatic axial disease in psoriatic spondyloarthropathy. A retrospective study Clin Rheumatol. 2002;21(1):10–3.
Jadon DR, Sengupta R, Nightingale A, Lindsay M, Korendowych E, Robinson G, et al. Axial disease in psoriatic arthritis study: defining the clinical and radiographic phenotype of psoriatic spondyloarthritis. Ann Rheum Dis. 2017;76(4):701–7.
Guttman-Yassky E, Del Duca E, Da Rosa J, Bar J, Ezzedine K, Ye Z, et al. Improvements in immune/melanocyte biomarkers with JAK3/TEC family kinase inhibitor ritlecitinib in vitiligo. J Allergy Clin Immunol. 2023. https://doi.org/10.1016/j.jaci.2023.09.021.
Nakayasu ES, Gritsenko M, Piehowski PD, Gao Y, Orton DJ, Schepmoes AA, et al. Tutorial: best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat Protoc. 2021;16(8):3737–60.
Greco V, Piras C, Pieroni L, Urbani A. Direct assessment of plasma/serum sample quality for proteomics biomarker investigation. Methods Mol Biol. 2017;1619:3–21.
Cui M, Cheng C, Zhang L. High-throughput proteomics: a methodological mini-review. Lab Invest. 2022;102(11):1170–81.
Petrera A, von Toerne C, Behler J, Huth C, Thorand B, Hilgendorff A, et al. Multiplatform approach for plasma proteomics: complementarity of olink proximity extension assay technology to mass spectrometry-based protein profiling. J Proteome Res. 2021;20(1):751–62.
Taylor W, Gladman D, Helliwell P, Marchesoni A, Mease P, Mielants H. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis Rheum. 2006;54(8):2665–73.
Ye L, Liu Y, **ao Q, Dong L, Wen C, Zhang Z, et al. MRI compared with low-dose CT scanning in the diagnosis of axial spondyloarthritis. Clin Rheumatol. 2020;39(4):1295–303.
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C. jvenn: an interactive venn diagram viewer. BMC Bioinform. 2014;15(1):293.
Michelena X, Poddubnyy D, Marzo-Ortega H. Axial psoriatic arthritis: a distinct clinical entity in search of a definition. Rheum Dis Clin N Am. 2020;46(2):327–41.
Proft F, Käding H. Same, same or different? commonalities and differences between spondyloarthritis and its subsets of axial and peripheral spondyloarthritis with psoriatic arthritis and its diverse phenotypes. RMD Open. 2023;9(2): e002872.
Mease P, van der Heijde D, Landewé R, Mpofu S, Rahman P, Tahir H, et al. Secukinumab improves active psoriatic arthritis symptoms and inhibits radiographic progression: primary results from the randomised, double-blind, phase III FUTURE 5 study. Ann Rheum Dis. 2018;77(6):890–7.
Poddubnyy D, Baraliakos X, Van den Bosch F, Braun J, Coates LC, Chandran V, et al. Axial involvement in psoriatic arthritis cohort (AXIS): the protocol of a joint project of the assessment of spondyloarthritis international society (ASAS) and the group for research and assessment of psoriasis and psoriatic arthritis (GRAPPA). Ther Adv Musculoskelet Dis. 2021. https://doi.org/10.1177/1759720X211057975.
Aydin SZ, Kilic L, Kucuksahin O, Ureyen SB, Kalyoncu U. Performances of inflammatory back pain criteria in axial psoriatic arthritis. Rheumatology. 2017;56(11):2031–2.
Diekhoff T, Eshed I, Radny F, Ziegeler K, Proft F, Greese J, et al. Choose wisely: imaging for diagnosis of axial spondyloarthritis. Ann Rheum Dis. 2022;81(2):237–42.
Tombran-Tink J, Johnson LV. Neuronal differentiation of retinoblastoma cells induced by medium conditioned by human RPE cells. Invest Ophthalmol Vis Sci. 1989;30(8):1700–7.
Filleur S, Nelius T, de Riese W, Kennedy RC. Characterization of PEDF: a multi-functional serpin family protein. J Cell Biochem. 2009;106(5):769–75.
Baxter-Holland M, Dass CR. Pigment epithelium-derived factor: a key mediator in bone homeostasis and potential for bone regenerative therapy. J Pharm Pharmacol. 2018;70(9):1127–38.
Hu K, Olsen BR. Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J Clin Invest. 2016;126(2):509–26.
Li F, Song N, Tombran-Tink J, Niyibizi C. Pigment epithelium-derived factor enhances differentiation and mineral deposition of human mesenchymal stem cells. Stem Cells. 2013;31(12):2714–23.
Burgers TA, Williams BO. Regulation of Wnt/β-catenin signaling within and from osteocytes. Bone. 2013;54(2):244–9.
Li F, Song N, Tombran-Tink J, Niyibizi C. Pigment epithelium derived factor suppresses expression of Sost/Sclerostin by osteocytes: implication for its role in bone matrix mineralization. J Cell Physiol. 2015;230(6):1243–9.
Akiyama T, Dass CR, Shinoda Y, Kawano H, Tanaka S, Choong PF. PEDF regulates osteoclasts via osteoprotegerin and RANKL. Biochem Biophys Res Commun. 2010;391(1):789–94.
Paine A, Ritchlin C. Altered bone remodeling in psoriatic disease: new insights and future directions. Calcif Tissue Int. 2018;102(5):559–74.
Wang F, Ma X, Zhou M, Pan X, Ni J, Gao M, et al. Serum pigment epithelium-derived factor levels are independently correlated with the presence of coronary artery disease. Cardiovasc Diabetol. 2013;12:56.
Cheng Q, **a W, Yang S, Ye P, Mei M, Song Y, et al. Association of serum pigment epithelium-derived factor with high-sensitivity C-reactive protein in women with polycystic ovary syndrome. J Endocrinol Invest. 2013;36(8):632–5.
Jenkins AJ, Zhang SX, Rowley KG, Karschimkus CS, Nelson CL, Chung JS, et al. Increased serum pigment epithelium-derived factor is associated with microvascular complications, vascular stiffness and inflammation in Type 1 diabetes. Diabet Med. 2007;24(12):1345–51.
Li X, Wang T, Yang T, Shen Y, An J, Liu L, et al. Elevated plasma levels of pigment epithelium-derived factor correlated with inflammation and lung function in COPD patients. Int J Chron Obstruct Pulmon Dis. 2015;10:587–94.
Hartl J, Kurth F, Kappert K, Horst D, Mülleder M, Hartmann G, et al. Quantitative protein biomarker panels: a path to improved clinical practice through proteomics. EMBO Mol Med. 2023;15(4): e16061.
Nakayasu ES, Gritsenko M, Piehowski PD, Gao Y, Orton DJ, Schepmoes AA, et al. Tutorial: best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat Protoc. 2021;16(8):3737–60.
Acknowledgements
We would like to express our gratitude to Bio Miao Biological Technology Co, Ltd (Bei**g, China) for their technical support.
Funding
This study was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1–005) and National High Level Hospital Clinical Research Funding (2022-PUMCH-B-013).
Author information
Authors and Affiliations
Contributions
CL, XL, and XZ contributed to the study’s conception and design; all authors contributed to material preparation, and data collection; CL contributed to data analysis. The first draft of the manuscript was written by CL and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Peking Union Medical College Hospital (K4397). Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1. Enrichment analysis of DEPs between axPsA and pPsA.(A) Gene Ontology (GO) classification of the DEPs. The top 10 enriched terms in the Biological Process (BP), Cellular Component (CC), and Molecular Function (MF) are listed. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the DEPs. The top 10 enriched pathways are listed. axPsA, axial psoriatic arthritis; pPsA, peripheral psoriatic arthritis; DEPs, differentially expressed proteins. Table S1. Multivariate logistic regression analysis for clinical characteristics and serum PEDF between axPsA and pPsA.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lu, C., Yang, F., He, S. et al. Serum proteome analysis identifies a potential biomarker for axial psoriatic arthritis. Eur J Med Res 29, 146 (2024). https://doi.org/10.1186/s40001-024-01731-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40001-024-01731-9