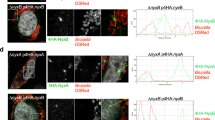
Abstract
Background
The TIR domain-containing proteins BtpA/Btp1/TcpB and BtpB are translocated into host cells by the facultative intracellular bacterial pathogen Brucella. Here, they interfere with Toll like receptor signalling to temper the host inflammatory response. BtpA has also been found to modulate microtubule dynamics. In both proteins we identified a WxxxE motif, previously shown to be an essential structural component in a family of bacterial type III secretion system effectors that modulate host actin dynamics by functioning as guanine nucleotide exchange factors of host GTPases. We analysed a role for the WxxxE motif in association of BtpA and BtpB with the cytoskeleton.
Results
Unlike BtpA, ectopically expressed BtpB did not show a tubular localisation, but was found ubiquitously in the cytoplasm and the nucleus, and often appeared in discrete punctae in HeLa cells. BtpB was able to protect microtubules from drug-induced destabilisation similar to BtpA. The WxxxE motif was important for the ability of BtpA and BtpB to protect microtubules against destabilising drugs. Surprisingly, ectopic expression of BtpA, although not BtpB, in HeLa cells induced the formation of filopodia. This process was invariably dependent of the WxxxE motif. Our recent resolution of the crystal structure of the BtpA TIR domain reveals that the motif positions a glycine residue that has previously been shown to be essential for interaction of BtpA with microtubules.
Conclusions
Our results suggest a structural role for the WxxxE motif in the association of BtpA and BtpB with microtubules, as with the WxxxE GEF family proteins where the motif positions an adjacent catalytic loop important for interaction with specific Rho GTPases. In addition, the ability of ectopically expressed BtpA to induce filopodia in a WxxxE-dependent manner suggests a novel property for BtpA. A conserved WxxxE motif is found in most bacterial and several eukaryotic TIR domain proteins. Despite the similarity between ectopically expressed BtpA and WxxxE GEFs to modulate host actin dynamics, our results suggest that BtpA is not part of this WxxxE GEF family. The WxxxE motif may therefore be a more common structural motif than thus far described. BtpA may provide clues to cross-talk between the TLR and GTPase signalling pathways.
Similar content being viewed by others
Background
Brucella is a Gram negative facultative intracellular bacterium that can cause brucellosis or Malta Fever, the world's most widespread zoonotic disease [1]. In humans Brucella causes an undulant fever that can be accompanied by complications such as endocarditis, arthritis and osteomyelitis [2]. Brucella is a stealth pathogen, known for its silent entry into host cells and multiple mechanisms to suppress host innate immune signalling, including non-toxic lipid A, and avoidance of oxidative burst and Toll like receptor (TLR) signalling cascades [2]-[6]. Whereas the primary task of immune cells is to phagocytose and degrade microbes, many intracellular pathogens, including Brucella, use secretion systems to introduce bacterial effector proteins directly into host cells to alter host cell biology and favour their intracellular replication [7],[8]. For full virulence, Brucella requires its VirB type IV secretion system (T4SS) to modulate endosomal trafficking, and create a replication niche in ER-derived membrane vesicles, named Brucella containing vacuoles (BCV) [9]-[11]. For many years, the effector proteins translocated by the Brucella VirB system remained elusive. After the identification of a first candidate [12], recent efforts from several laboratories, using both bioinformatics screens and translocation assays have resulted in a list of possible effectors [13]-[16]. To date, the precise role of most of these proteins in Brucella virulence is still not clear and the subject of intense research.
Recently, the proteins BtpA (Btp1/TcpB) and BtpB have been shown to be translocated by Brucella into host cells [17]. Although a TEM-1 β-lactamase assay did not show a significant difference in translocation efficiency of the proteins from wild type and virB mutant bacteria, a CyA reporter assay showed VirB-dependent protein transport into host cells, suggesting BtpA and BtpB may be substrates of the VirB T4SS. BtpA and BtpB belong to a class of bacterial proteins first described in Salmonella, Escherichia coli and Brucella that share homology with the eukaryotic Toll/Interleukin-1 receptor (TIR) domain [18],[19]. A conserved TIR domain is present in eukaryotic TLR proteins as well as their downstream signalling TIR adaptor proteins, including the central cytosolic adapter protein MyD88. The TIR domain is essential for TLR and adaptor interactions and for the onset of a signalling cascade resulting in nuclear translocation of the transcription factor NFκB, followed by the production of pro-inflammatory cytokines and type I interferons [20]. TIR domain interactions play a key role in activating conserved cellular signal transduction pathways in response to pathogen signals, and it was suggested that bacterial TIR proteins interfere with host TLR defence signalling by molecular mimicry (reviewed in [21]).
B. abortus 2308 BtpA (BAB1_0279) and the almost identical B. melitensis 16M BtpA (BMEI1674) down modulate maturation of dendritic cells [22] and inhibit TLR-induced NFκB activation. It has been suggested that this is through interference with the TLR4/MyD88/TIRAP complex [23]-[27], however the exact binding partner of BtpA is still a subject of controversy. Far less is known about BtpB, however recently it has also been shown to play a role in immune modulation [17]. Recently we, and others, published the crystal structure of the BtpA TIR domain, which showed a dimeric arrangement of a canonical TIR domain [25],[28],[ For cloning Escherichia coli DH5α was cultured at 37°C in Luria-Bertani broth (LB) (Invitrogen, Merck). Kanamycin (25 μg/ml) or chloramphenicol (30 μg/ml) were added to the media when appropriate. A 828 bp version of the btpA gene was cloned from B. melitensis 16M genomic DNA (BMEI1674, Genbank accession NP_540591 is the shorter annotated version) using PCR (primers Bgl II L1674-F 5'-GAAGATCT TATGAGTTCGTACTCTTCTAATATTG-3', and Pst IL1674-R 5'-AACTGCAG TCAGATAAGGGAATGCAGTTC-3') and cloned in frame with the GFP coding sequence in eukaryotic expression plasmid pEGFP (Clontech) using standard protocols, resulting in plasmid pIN271. A 978 version of the btpB gene was cloned from B. suis genomic DNA (BR0735, Genbank accession number AAN29664 is the shorter annotated version) using PCR (primers Bgl II 735-F 5'-GAAGATCT TATGACATCTAGTCGCGACACG-3', and Pst I 735-R 5'-AACTGCAG CTAGGTGATGAGGGCGACG-3') and cloned in frame with the GFP coding sequence in pEGFP resulting in plasmid pIN292. We amplified a longer version of btpB, which was recently also shown to complement a B. abortus btpB mutant in the control of NF-κB translocation into the nucleus [17]. Site directed mutagenesis of the WxxxE motif was done using the QuickChange Site-Directed Mutagenesis kit (Stratagene) following manufacturer's instructions, using pIN271 as a template with the following mutagenic primers for btpA (changed codons for Trp (TGG) and Glu (GAA) are italicised): W213A (5'-TTTAGCAAGCAAGCG CCCGCAAGAGAATTAG-3'), W213S (5'-TTTAGCAAGCAATCG CCCGCAAGAGAATTAG-3'), W213F (5'-TTTAGCAAGCAATTC CCCGCAAGAGAATTAG-3'), E217A (5'-CAATGGCCCGCAAGAGCA TTAGATGGACTGAC-3'), E217D (5'- CAATGGCCCGCAAGAGAT TTAGATGGACTGAC-3'), I226S (5'- CTGACGGCAATGGAAAGT GGCGGACAGACGC-3'), G183A (5'- CATATACGTTGAAGGTCGCT GACAGCCTTCGGCG-3'), and btpB (pIN292 as a template): W263S (5'- CTATCAGCGAAAAGACTCG TGCGGCGTCG-3') and E267A (5'- CTGGTGCGGCGTCGCG TTCCGCGCGATTCG-3'). Constructs were verified by DNA sequencing (Eurofins MWG operon, Germany). Table 2 summarizes the plasmids used in this study. HeLa cells were grown in RPMI 1640 (Gibco) supplemented with heat-inactivated 10% fetal bovine serum (FBS, Lonza, Switzerland). Transfections were performed using Lipofectamine 2000 (Invitrogen). For immuno fluorescence studies, cells were seeded on coverslips (BD Bioscience) and cultured overnight in 12-well dishes. Cells were transfected and, after 16–20 hours, treated with nocodazole (Sigma M1404) at a concentration of 1 μg/ml for 30 min if desired. Cells were fixed with 4% PFA, and processed for immunocytochemistry. Monoclonal mouse anti-β-tubulin (Sigma Aldrich, T4026), mouse anti-FK2 (Enzo Life science, BML-PW8810), Texas Red anti-mouse (Vector Laboratories, TI-2000), Rhodamin phalloidin (Invitrogen R415), Rabbit anti-BtpA (a gift from Marty Roop, East Carolina University, Greenville), FITC anti-rabbit (Vector Laboratories, FI-2000) were used for immune labelling. Immuno fluorescence microscopy was performed using a LEICA DM/IRB microscope using filter sets L5 (band pass (BP) 480/40; Beam splitter (BS) 505; emission BP527/30) and N2.1 (515–560; BS 580; emission long pass (LP) 590), respectively. For imaging we used a Coolsnap fx (Roper Scientifique) and MetaVue software, and images were further processed using Adobe Photoshop. Confocal analysis was performed at the RIO imaging platform in Montpellier, with a Biorad MRC1024 confocal microscope. Brucella genomes were obtained and compared on the PATRIC website [33]. Sequence alignments were performed using T-Coffee analysis, and further analysed using Jalview. Structural figures were generated with Pymol (www.pymol.org).Materials and methods
Bacterial strains
Plasmids and site-directed mutagenesis
Cell culture, transfection, immunocytochemistry, and fluorescence microscopy
Bio informatic analysis
Additional files
Abbreviations
- GEF:
-
Guanidine exchange factors
- LPS:
-
Lipopolysaccharide
- MTOC:
-
Microtubule-organizing centre
- NC:
-
Nocodazole
- TIR:
-
Toll/Interleukin-1 receptor
- TLR:
-
Toll like receptor
- T4SS:
-
Type IV secretion system
- T3SS:
-
Type III secretion system
- UPS:
-
Ubiquitin Proteasome system
References
O’Callaghan D, Whatmore AM: Brucella genomics as we enter the multi-genome era. Brief Funct Genomics. 2011, 10: 334-341. 10.1093/bfgp/elr026.
Martirosyan A, Moreno E, Gorvel J-P: An evolutionary strategy for a stealthy intracellular Brucella pathogen. Immunol Rev. 2011, 240: 211-234. 10.1111/j.1600-065X.2010.00982.x.
Cardoso PG, Macedo GC, Azevedo V, Oliveira SC: Brucella spp noncanonical LPS: structure, biosynthesis, and interaction with host immune system. Microb Cell Fact. 2006, 5: 13-10.1186/1475-2859-5-13.
Celli J: Surviving inside a macrophage: the many ways of Brucella. Res Microbiol. 2006, 157: 93-98. 10.1016/j.resmic.2005.10.002.
De Jong MF, Rolán HG, Tsolis RM: Innate immune encounters of the (Type) 4th kind: Brucella. Cell Microbiol. 2010, 12: 1195-1202. 10.1111/j.1462-5822.2010.01498.x.
Seleem MN, Boyle SM, Sriranganathan N: Brucella: a pathogen without classic virulence genes. Vet Microbiol. 2008, 129: 1-14. 10.1016/j.vetmic.2007.11.023.
Llosa M, Roy C, Dehio C: Bacterial type IV secretion systems in human disease. Mol Microbiol. 2009, 73: 141-151. 10.1111/j.1365-2958.2009.06751.x.
Bhatty M, Laverde Gomez JA, Christie PJ: The expanding bacterial type IV secretion lexicon. Res Microbiol. 2013, 164: 620-639. 10.1016/j.resmic.2013.03.012.
Von Bargen K, Gorvel J-P, Salcedo SP: Internal affairs: investigating the Brucella intracellular lifestyle. FEMS Microbiol Rev. 2012, 36: 533-562. 10.1111/j.1574-6976.2012.00334.x.
O’ Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M: A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol. 1999, 33: 1210-1220. 10.1046/j.1365-2958.1999.01569.x.
Starr T, Ng TW, Wehrly TD, Knodler LA, Celli J: Brucella intracellular replication requires trafficking through the late endosomal/lysosomal compartment. Traffic. 2008, 9: 678-694. 10.1111/j.1600-0854.2008.00718.x.
Lavigne J-P, Patey G, Sangari FJ, Bourg G, Ramuz M, O’Callaghan D, Michaux-Charachon S: Identification of a New Virulence Factor, BvfA, in Brucella suis. Infect Immun. 2005, 73: 5524-5529. 10.1128/IAI.73.9.5524-5529.2005.
De Jong MF, Sun Y-H, den Hartigh AB, van Dijl JM, Tsolis RM: Identification of VceA and VceC, two members of the VjbR regulon that are translocated into macrophages by the Brucella type IV secretion system. Mol Microbiol. 2008, 70: 1378-1396. 10.1111/j.1365-2958.2008.06487.x.
De Barsy M, Jamet A, Filopon D, Nicolas C, Laloux G, Rual J-F, Muller A, Twizere J-C, Nkengfac B, Vandenhaute J, Hill DE, Salcedo SP, Gorvel J-P, Letesson J-J, De Bolle X: Identification of a Brucella spp. secreted effector specifically interacting with human small GTPase Rab2. Cell Microbiol. 2011, 13: 1044-1058. 10.1111/j.1462-5822.2011.01601.x.
Marchesini MI, Herrmann CK, Salcedo SP, Gorvel J-P, Comerci DJ: In search of Brucella abortus type IV secretion substrates: screening and identification of four proteins translocated into host cells through VirB system. Cell Microbiol. 2011, 13: 1261-1274. 10.1111/j.1462-5822.2011.01618.x.
Myeni S, Child R, Ng TW, Kupko JJ, Wehrly TD, Porcella SF, Knodler LA, Celli J: Brucella Modulates Secretory Trafficking via Multiple Type IV Secretion Effector Proteins. PLoS Pathog. 2013, 9: e1003556-10.1371/journal.ppat.1003556.
Salcedo SP, Marchesini MI, Degos C, Terwagne M, Von Bargen K, Lepidi H, Herrmann CK, Santos Lacerda TL, Imbert PRC, Pierre P, Alexopoulou L, Letesson JJ, Comerci DJ, Gorvel JP: BtpB, a novel Brucella TIR-containing effector protein with immune modulatory functions. Front Cell Infect Microbiol. 2013, 3: 28-10.3389/fcimb.2013.00028.
Newman RM, Salunkhe P, Godzik A, Reed JC: Identification and Characterization of a Novel Bacterial Virulence Factor That Shares Homology with Mammalian Toll/Interleukin-1 Receptor Family Proteins. Infect Immun. 2006, 74: 594-601. 10.1128/IAI.74.1.594-601.2006.
Cirl C, Wieser A, Yadav M, Duerr S, Schubert S, Fischer H, Stappert D, Wantia N, Rodriguez N, Wagner H, Svanborg C, Miethke T: Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat Med. 2008, 14: 399-406. 10.1038/nm1734.
Kawai T, Akira S: Toll-like Receptors and Their Crosstalk with Other Innate Receptors in Infection and Immunity. Immunity. 2011, 34: 637-650. 10.1016/j.immuni.2011.05.006.
Rana RR, Zhang M, Spear AM, Atkins HS, Byrne B: Bacterial TIR-containing proteins and host innate immune system evasion. Med Microbiol Immunol. 2013, 202: 1-10. 10.1007/s00430-012-0253-2.
Salcedo SP, Marchesini MI, Lelouard H, Fugier E, Jolly G, Balor S, Muller A, Lapaque N, Demaria O, Alexopoulou L, Comerci DJ, Ugalde RA, Pierre P, Gorvel J-P: Brucella Control of Dendritic Cell Maturation Is Dependent on the TIR-Containing Protein Btp1. PLoS Pathog. 2008, 4: e21-10.1371/journal.ppat.0040021.
Radhakrishnan GK, Splitter GA: Biochemical and functional analysis of TIR domain containing protein from Brucella melitensis. Biochem Biophys Res Commun. 2010, 397: 59-63. 10.1016/j.bbrc.2010.05.056.
Radhakrishnan GK, Yu Q, Harms JS, Splitter GA: Brucella TIR Domain-containing Protein Mimics Properties of the Toll-like Receptor Adaptor Protein TIRAP. J Biol Chem. 2009, 284: 9892-9898. 10.1074/jbc.M805458200.
Alaidarous M, Ve T, Casey LW, Valkov E, Ericsson DJ, Ullah MO, Schembri MA, Mansell A, Sweet MJ, Kobe B: Mechanism of bacterial interference with TLR4 signaling by Brucella TIR-domain-containing protein TcpB. J Biol Chem. 2013, 289: 654-668. 10.1074/jbc.M113.523274.
Chaudhary A, Ganguly K, Cabantous S, Waldo GS, Micheva-Viteva SN, Nag K, Hlavacek WS, Tung C-S: The Brucella TIR-like protein TcpB interacts with the death domain of MyD88. Biochem Biophys Res Commun. 2012, 417: 299-304. 10.1016/j.bbrc.2011.11.104.
Sengupta D, Koblansky A, Gaines J, Brown T, West AP, Zhang D, Nishikawa T, Park S-G, Roop RM, Ghosh S: Subversion of innate immune responses by Brucella through the targeted degradation of the TLR signaling adapter, MAL. J Immunol. 2010, 184: 956-964. 10.4049/jimmunol.0902008.
Kaplan-Türköz B, Koelblen T, Felix C, Candusso M-P, O’Callaghan D, Vergunst AC, Terradot L: Structure of the Toll/interleukin 1 receptor (TIR) domain of the immunosuppressive Brucella effector BtpA/Btp1/TcpB. FEBS Lett. 2013, 587: 3412-3416. 10.1016/j.febslet.2013.09.007.
Snyder GA, Deredge D, Waldhuber A, Fresquez T, Wilkins DZ, Smith PT, Duerr S, Cirl C, Jiang J, Jennings W, Luchetti T, Snyder N, Sundberg EJ, Wintrode P, Miethke T, **ao TS: Crystal structures of the TIR domains from the Brucella protein TcpB and host adapter TIRAP reveal mechanisms of molecular mimicry. J Biol Chem. 2014, 289: 669-679. 10.1074/jbc.M113.523407.
Radhakrishnan GK, Harms JS, Splitter GA: Modulation of microtubule dynamics by a TIR domain protein from the intracellular pathogen Brucella melitensis. Biochem J. 2011, 439: 79-83. 10.1042/BJ20110577.
Orchard RC, Alto NM: Mimicking GEFs: A Common Theme For Bacterial Pathogens. Cell Microbiol. 2012, 14: 10-18. 10.1111/j.1462-5822.2011.01703.x.
Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, Dixon JE: Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006, 124: 133-145. 10.1016/j.cell.2005.10.031.
Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, Machi D, Mao C, Nordberg EK, Olson R, Overbeek R, Pusch GD, Shukla M, Schulman J, Stevens RL, Sullivan DE, Vonstein V, Warren A, Will R, Wilson MJC, Yoo HS, Zhang C, Zhang Y, Sobral BW: PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42: D581-D591. 10.1093/nar/gkt1099.
Huang Z, Sutton SE, Wallenfang AJ, Orchard RC, Wu X, Feng Y, Chai J, Alto NM: Structural insights into host GTPase isoform selection by a family of bacterial GEF mimics. Nat Struct Mol Biol. 2009, 16: 853-860. 10.1038/nsmb.1647.
Zhang Q, Zmasek CM, Cai X, Godzik A: TIR domain-containing adaptor SARM is a late addition to the ongoing microbe-host dialog. Dev Comp Immunol. 2011, 35: 461-468. 10.1016/j.dci.2010.11.013.
Carty M, Goodbody R, Schröder M, Stack J, Moynagh PN, Bowie AG: The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat Immunol. 2006, 7: 1074-1081. 10.1038/ni1382.
Bulgin R, Raymond B, Garnett JA, Frankel G, Crepin VF, Berger CN, Arbeloa A: Bacterial guanine nucleotide exchange factors SopE-like and WxxxE effectors. Infect Immun. 2010, 78: 1417-1425. 10.1128/IAI.01250-09.
Chen C-T, Ettinger AW, Huttner WB, Doxsey SJ: Resurrecting remnants: the lives of post-mitotic midbodies. Trends Cell Biol. 2013, 23: 118-128. 10.1016/j.tcb.2012.10.012.
Mukai A, Mizuno E, Kobayashi K, Matsumoto M, Nakayama KI, Kitamura N, Komada M: Dynamic regulation of ubiquitylation and deubiquitylation at the central spindle during cytokinesis. J Cell Sci. 2008, 121: 1325-1333. 10.1242/jcs.027417.
Ben Tekaya H, Gorvel JP, Dehio C: Bartonella and Brucella--weapons and strategies for stealth attack. Cold Spring Harb Perspect Med. 2013, 3. doi:10.1101/cshperspect.a010231
Fekonja O, Benčina M, Jerala R: Toll/interleukin-1 receptor domain dimers as the platform for activation and enhanced inhibition of Toll-like receptor signaling. J Biol Chem. 2012, 287: 30993-31002. 10.1074/jbc.M112.376186.
Smith JA, Khan M, Magnani DD, Harms JS, Durward M, Radhakrishnan GK, Liu Y-P, Splitter GA: Brucella Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages. PLoS Pathog. 2013, 9: e1003785-10.1371/journal.ppat.1003785.
Hanson PI, Cashikar A: Multivesicular body morphogenesis. Annu Rev Cell Dev Biol. 2012, 28: 337-362. 10.1146/annurev-cellbio-092910-154152.
Lafarga M, Berciano MT, Pena E, Mayo I, Castan G, Bohmann J, Pedro Rodrigues D, Paulo J, Tavanez J, Carmo Fonseca M: Clastosome: A Subtype of Nuclear Body Enriched in 19S and 20S Proteasomes, Ubiquitin, and Protein Substrates of Proteasome. Mol Biol Cell. 2002, 13: 2771-2782. 10.1091/mbc.E02-03-0122.
Kopito RR: Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000, 68: 524-530. 10.1016/S0962-8924(00)01852-3.
Angot A, Vergunst A, Genin S, Peeters N: Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 2007, 3: e3-10.1371/journal.ppat.0030003.
Ashida H, Kim M, Sasakawa C: Exploitation of the host ubiquitin system by human bacterial pathogens. Nat Rev Microbiol. 2014, 12: 399-413. 10.1038/nrmicro3259.
Radhakrishnan GK, Spliiter GA: Modulation of host microtubule dynamics by pathogenic bacteria. Biomol Concepts. 2012, 3: 571-580. 10.1515/bmc-2012-0030.
Aktories K: Bacterial protein toxins that modify host regulatory GTPases. Nat Rev Microbiol. 2011, 9: 487-498. 10.1038/nrmicro2592.
Kenny B, Ellis S, Leard AD, Warawa J, Mellor H, Jepson MA: Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol Microbiol. 2002, 44: 1095-1107. 10.1046/j.1365-2958.2002.02952.x.
Arbeloa A, Bulgin RR, MacKenzie G, Shaw RK, Pallen MJ, Crepin VF, Berger CN, Frankel G: Subversion of actin dynamics by EspM effectors of attaching and effacing bacterial pathogens. Cell Microbiol. 2008, 10: 1429-1441. 10.1111/j.1462-5822.2008.01136.x.
Berger CN, Crepin VF, Jepson MA, Arbeloa A, Frankel G: The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cell Microbiol. 2009, 11: 309-322. 10.1111/j.1462-5822.2008.01254.x.
Cirl C, Miethke T: Microbial Toll/interleukin 1 receptor proteins: a new class of virulence factors. Int J Med Microbiol. 2010, 300: 396-401. 10.1016/j.ijmm.2010.04.001.
Truttmann MC, Guye P, Dehio C: BID-F1 and BID-F2 Domains of Bartonella henselae Effector Protein BepF Trigger Together with BepC the Formation of Invasome Structures. PLoS ONE. 2011, 6: e25106-10.1371/journal.pone.0025106.
Arbibe L, Mira JP, Teusch N, Kline L, Guha M, Mackman N, Godowski PJ, Ulevitch RJ, Knaus UG: Toll-like receptor 2-mediated NF-kappa B activation requires a Rac1-dependent pathway. Nat Immunol. 2000, 1: 533-540. 10.1038/82797.
Van Acker T, Eyckerman S, Vande Walle L, Gerlo S, Goethals M, Lamkanfi M, Bovijn C, Tavernier J, Peelman F: The Small GTPase Arf6 Is Essential for the Tram/Trif Pathway in TLR4 Signaling. J Biol Chem. 2014, 289: 1364-1376. 10.1074/jbc.M113.499194.
Uronen-Hansson H, Allen J, Osman M, Squires G, Klein N, Callard RE: Toll-like receptor 2 ( TLR2) and TLR4 are present inside human dendritic cells, associated with microtubules and the Golgi apparatus but are not detectable on the cell surface : integrity of microtubules is required for interleukin-12 production in resp. Immunology. 2004, 111: 173-178. 10.1111/j.0019-2805.2003.01803.x.
Acknowledgements
The authors thank Marty Roop for BtpA antibodies, and Philippe Fort and Géraldine Pawlak for helpful discussions. We thank Montpellier RIO imaging for confocal microscopy. U1047 received funding from INSERM, Université Montpellier 1, the Region Languedoc-Roussillon, the Agence Nationale de Recherche (ANR-MIME; T4SS, ANR-MIE; BruCell). Work in the LT group was funded by the ATIP-Avenir program from the CNRS and Ligue Contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
Conceived and designed the experiments: CF, LT, DOC, ACV. Performed the experiments: CF, BKT, TK, SR. Analysed the data: CF. SR, BKT, LT, DOC, ACV. Contributed to writing the paper: CF, LT, DOC, ACV. All authors read and approved the final manuscript.
Electronic supplementary material
12964_2014_53_MOESM1_ESM.pdf
Additional file 1: Figure S1.: Evolutionary tree of Brucella species and distribution of BtpA and BtpB proteins. The alignment was made in Patric and the tree was optimized using Mega Software. btpA and btpB genes are highly conserved throughout the genus. We found only four variations in 407 sequences (showing 47 species). All B. melitensis strains carry a single amino acid change (leading to A163V), while three strains carry a variation in R167H (B. ovis), G200D (B. suis ATCC23445) or V182G (B. neotomae). BtpB is also highly conserved, with only two amino acids that show variation. The Trp residue, as part of a WxxxE motif, in BtpB was replaced by an Arg residue in B. melitensis BtpB (W263R). (PDF 492 KB)
12964_2014_53_MOESM2_ESM.pdf
Additional file 2: Figure S2.: Confocal imaging of BtpA in HeLa cells shows tubular localisation at cell periphery. HeLa cells were transiently transfected with a plasmid encoding GFP-BtpA, fixed after 16 h and analysed by confocal microscopy. The coloured image (green) shows the merged stack images of 14 Z-slices, the other images show the individual Z-images (1 μm depth) of HeLa cells expressing GFP-BtpA. Bar, 25 μm. (PDF 277 KB)
12964_2014_53_MOESM3_ESM.pdf
Additional file 3: Figure S3.: Ectopic expression of BtpA in HeLa cells shows colocalisation with microtubules HeLa cells were transiently transfected with a plasmid expressing GFP-BtpA. At 16 h cells were fixed and processed for immuno labelling with mouse anti β-tubulin antibody, detected with anti-mouse Texas Red to visualize microtubules (red fluorescence). The large image shows a merged image of 6 Z-Stack confocal images (1 μm depth) of cells with microtubules (in red) and GFP-BtpA in one of the cells. Arrow heads indicate colocalisation of GFP-BtpA with the microtubule network (yellow), at the periphery of the cell. The other images represent the individual Z-stack images. Scale bar, 25 μm. Data are representative of 3 or more independent experiments. (PDF 309 KB)
12964_2014_53_MOESM4_ESM.pdf
Additional file 4: Figure S4.: Expression phenotypes of GFP-BtpB in HeLa cells. HeLa cells were transiently transfected with a plasmid expressing GFP-BtpB. At 16 h cells were fixed, processed, and analysed using fluorescence microscopy. The individual images show the different observed expression patterns ranging from a diffuse signal, to cells with a diffuse fluorescent signal as well as accumulation of BtpB in punctae, heterogeneous in size and number. Scale bar, 25 μm. (PDF 192 KB)
12964_2014_53_MOESM5_ESM.pdf
Additional file 5: Figure S5.: Colocalisation of BtpB and conjugated ubiquitin. HeLa cells were transiently transfected with a plasmid expressing GFP-BtpB. At 16 h cells were fixed and processed for immune labelling with mAb FK2, which detects mono- and poly conjugated ubiquitin, and labelled with anti-mouse Texas Red for fluorescence detection. A. Confocal images showing stacks of overlay and individual GFP-BtpB and FK2 images. Scale bar, 10 μm. B. Two fluorescence images (red and green filters) showing colocalisation of BtpB and FK2, also at the intercellular bridge in late cytokinesis (arrow, and boxed areas). Scale bar, 25 μm C. Image showing transfected and non-transfected cells with and without FK2 positive foci. Cells indicated with numbers in the overlay panel (on the right) are enlarged below as individual green and red fluorescent images. Scale bar, 25 μm. 1, 2. Transfected cells with a punctate pattern for BtpB, colocalising with FK2. FK2 positive, BtpB negative foci are also observed. 3, 4: Non-transfected, FK2 positive foci. 5. Transfected, no visible BtpB punctae, FK2 positive. (PDF 805 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Felix, C., Kaplan Türköz, B., Ranaldi, S. et al. The Brucella TIR domain containing proteins BtpA and BtpB have a structural WxxxE motif important for protection against microtubule depolymerisation. Cell Commun Signal 12, 53 (2014). https://doi.org/10.1186/s12964-014-0053-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12964-014-0053-y