Abstract
Background
Tetramethylpyrazine (TTMP) is a flavoring additive that significantly contributes to the formation of flavor compounds in soybean-based fermented foods. Over recent years, the application of TTMP in the food industry and medicine has been widely investigated. In addition, several methods for the industrial-scale production of TTMP, including chemical and biological synthesis, have been proposed. However, there have been few reports on the synthesis of TTMP through amino acid metabolic flux. In this study, we investigated genetic alterations of arginine metabolic flux in solid-state fermentation (SSF) of soybeans with Bacillus subtilis (B.subtilis) BJ3-2 to enhance the TTMP yield.
Results
SSF of soybeans with BJ3-2 exhibited a strong Chi-flavour (a special flavour of ammonia-containing smelly distinct from natto) at 37 °C and a prominent soy sauce-like aroma at 45 °C. Transcriptome sequencing and RT-qPCR verification showed that the rocF gene was highly expressed at 45 °C but not at 37 °C. Moreover, the fermented soybeans with BJ3-2ΔrocF (a rocF knockout strain in B. subtilis BJ3-2 were obtained by homologous recombination) at 45 °C for 72 h displayed a lighter color and a slightly decreased pH, while exhibiting a higher arginine content (increased by 14%) than that of BJ3-2. However, the ammonia content of fermented soybeans with BJ3-2ΔrocF was 43% lower than that of BJ3-2. Inversely, the NH4+ content in fermented soybeans with BJ3-2ΔrocF was increased by 28% (0.410 mg/kg). Notably, the TTMP content in fermented soybeans with BJ3-2ΔrocF and BJ3-2ΔrocF + Arg (treated with 0.05% arginine) were significantly increased by 8.6% (0.4617 mg/g) and 18.58% (0.504 mg/g) respectively than that of the BJ3-2.
Conclusion
The present study provides valuable information for understanding the underlying mechanism during the TTMP formation process through arginine metabolic flux.
Similar content being viewed by others
Background
Tetramethylpyrazine (TTMP) is a nitrogen-containing heterocyclic compound that contributes to the formation of many aromas and flavors compounds [10, 11]. The biological synthesis of TTMP provides several advantages over chemical syntheses, considering it is an environmentally friendly and cost-effective process [5]. Moreover, pyrazines have been widely reported as important microbial secondary metabolite, which implies that microbial metabolism can produce the TTMP [12].
Fermented soybeans (also named Douchi) are one of the most popular foods in China due to the strong Chi-flavour (namely soybean-flavour [13], a special flavor of ammonia-containing smelly distinct from natto). Bacillus subtilis (B. subtilis) is one of the most important microorganisms for the fermentation process of Douchi. More than half-century ago, Kosuge et al. [14] first suggested that B. subtilis can synthesize TTMP. Moreover, a recent study proved that TTMP in Chinese liquor is mainly generated from B. subtilis under Micro-Oxygen conditions [6]. Thus, it has been proposed that the metabolic engineering of B. subtilis can enhance the production of TTMP. A high yield (2.5 g/kg) of TTMP was obtained using B. subtilis IFO 3013 inoculated soybeans and fermentation for 14 days [15]. However, industrial production still has several difficulties, such as low production [16] and a low conversion rate of precursor substances [17].
Over the years, genetic engineering has emerged as a powerful tool in studying the behavior of the production of TTMP [7, 18]. Meng et al. [5] showed that knocking the 2,3-butanediol dehydrogenase gene (bdhA) and adding 2,3-butanediol exogenously improve the production of TTMP in B. subtilis. Similarly, a mutant bdhA and glucose uptake protein (GlcU) were reported to affect the production of TTMP in B. subtilis 168 [6]. Moreover, two genes (BDH1 and another BDH2 coding 2,3-butanediol dehydrogenase) were deleted or overexpressed to improve the TTMP yield in Saccharomyces cerevisiae [19]. The TTMP can be generated from various precursors, including acetoin and ammonia [20]. Alterations of carbon flux into the acetoin biosynthesis pathway by blocking the degradation and competing pathways can enhance the TTMP yield [5]. Moreover, the TTMP has also been produced by condensing acetoin with NH4+ [21, 22]. However, there have been few reports on the synthesis of TTMP through amino acid metabolic flux. Recently, a new theory has been proposed suggesting that amino acids have close relationship with the production of volatile compounds such as pyrazines [23]. Various metabolites were generated by using B. subtilis during the fermentation of natto, such as peptone, peptides, amino acids, sugars, and organic acids, enhancing organoleptic of the final products [24]. Yet, their contribution to the organoleptic properties of the final products has not yet been investigated in detail.
In this study, we investigated genetic alterations of arginine metabolic flux in order to enhance the TTMP yield. The results showed that the fermented soybeans with BJ3-2ΔrocF had a significantly higher content of arginine and TTMP. Our findings provided detailed insights into the effects of rocF genes on the metabolism of arginine and TTMP in fermented soybeans with the B. subtilis BJ3-2.
Results
Summary of the sequencing data
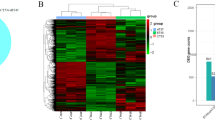
The sensory evaluation of the fermented broth was performed by well-trained panelists on the three components that included soy sauce-like aroma, Chi-flavour, and ammonia; individual scores were added together to provide a total score (Table 1). According to the sensory evaluation, we found that the Chi-flavour of broth fermented with BJ3-2 at 37 °C was prominent, whereas the soy sauce-like aroma of broth fermented with BJ3-2 at 45 °C was prominent; the ammonia was similar at 37 °C and 45 °C (Table 1). Therefore, we performed RNA sequencing (RNA-seq) analysis of BJ3-2 at 37 °C and 45 °C. For the sequencing data, we found that the base content was evenly distributed, and the nucleotide distributions were within a reasonable range (Additional file 1: Fig. S1A, D). The quality and error rate of the samples were within a normal range (Additional file 1: Fig. S1B, C, E and F). After filtering the original data, checking the sequencing error rate, and verifying the distribution of the GC content, clean reads for follow-up analysis were also obtained. The Q20 and Q30 values of BJ3-2 at 37 °C were 98.5% and 95.93%, respectively, while they were 98.42% and 95.7% at 45 °C (Table 2). Next, gene expression analysis indicated that the gene expression levels in different samples were uniformly distributed in density diagrams (Additional file 1: Fig. S2). After normalizing the sequencing data, we constructed a scatter and volcano diagram (Fig. 1A, B). As shown, 3648 genes were identified, including 67 up-regulated DEGs and 56 down-regulated DEGs (P-value < 0.05 and |log2FC|≥ 1) (Table 3).
GO and KEGG enrichment analysis of the DEGs
To determine the functions of the DEGs, GO analysis revealed that these genes were based on their roles in biological processes (BPs), cellular components (CCs), and molecular functions (MFs). The results indicated that these DEGs were mainly enriched in 28 GO terms (11 BPs, 10 CCs, and 7 MFs; Q-value ≤ 1). For BPs, the exhaustive analysis showed that many DEGs were involved in cellular processes, metabolic processes, and single-organism processes. For CCs, most DEGs were enriched in the cell, cell part, and macromolecular complex. For MFs, many DEGs were involved in catalytic activity, binding, and transporter activity (Fig. 1C). Additionally, KEGG pathway analysis revealed that the dominant DEGs were enriched in pyrimidine metabolism and arginine and proline metabolism (Fig. 1D; Additional file 2: Table S4). Collectively, the results suggested that the expression of genes related to various pathways differed in BJ3-2 cultured at 37 °C and 45 °C.
The expression of rocF in BJ3-2 incubated at 37 °C and 45 °C
Through the analysis of RNA-seq data from BJ3-2 incubated at different temperatures, we observed that several genes were involved in arginine and proline metabolism (Table 4), which are closely associated with the products of TTMP in B. subtilis [21], thus suggesting that these genes may affect the products of TTMP. Among them, the expression of three genes (fadM, argF, and argD) was up-regulated at least 1 ~ fold (log2FC), and five genes (rocD, rocF, sped, rocG, and rocA) was down-regulated at least 3 ~ fold (log2FC) after incubation at 45 °C. Hampel et al. suggested that the knockdown of rocD affects the normal growth of microorganisms [25]. Interestingly, the expression of rocF was down-regulated by 4.16 ~ fold (log2FC) after incubation at 45 °C (Table 4). Therefore, we selected rocF for further investigation and its expression in BJ3-2 incubated at different temperatures by RT-qPCR. The results showed that the dissolution curve of rocF was a single peak, and the amplification curve had a high degree of coincidence (Additional file 1: Fig. S3). The expression of rocF was low at 45 °C and high at 37 °C. Notably, the RT-qPCR results were consistent with the RNA-seq data (Fig. 2).
Generation of the rocF knockout strain
To reveal the role of the rocF gene, the knockout vector pUC18-HLarm-cm-HRarm of rocF was constructed and transformed into B. subtilis BJ3-2 (Additional file 1: Fig. S4). The positive clones were verified by PCR (Additional file 1: Fig. S4E). Subsequently, the positive clones were sequenced, and the results indicated that the rocF were successfully replaced by a cm fragment in the pUC18-HLarm-cm-HRarm vector (Additional file 1: Fig. S5). These results demonstrated that rocF of B. subtilis BJ3-2 was successfully knocked out and denoted by BJ3-2ΔrocF.
Characterization of the BJ3-2ΔrocF strain
To investigate the growth ability of BJ3-2ΔrocF, the growth characteristics of BJ3-2ΔrocF were assessed. As expected, the color and shape (rough surface, irregular shape, and surrounding folds) of BJ3-2ΔrocF on the plate were similar to BJ3-2 (Fig. 3A, B). In addition, the microscopic morphology of BJ3-2ΔrocF was short and rod-shaped, similar to BJ3-2. Moreover, BJ3-2ΔrocF was also purple, similar to BJ3-2, as shown by Gram staining (Fig. 3C, D). The results indicated no significant differences in the morphology and color by Gram staining were observed between BJ3-2ΔrocF and BJ3-2. Correspondingly, almost the same results were observed for the growth curves of BJ3-2ΔrocF and BJ3-2 (Fig. 3E). However, the arginase activity was significantly inhibited in BJ3-2ΔrocF compared to BJ3-2 (Fig. 4). These results suggest that knockout of rocF only contributes to the inhibition of the arginase activity, but not change the growth rate of BJ3-2.
Sensory evaluation of fermented soybeans
To assess the fermentation characteristics of BJ3-2ΔrocF in soybeans, the BJ3-2ΔrocF and BJ3-2 strains were inoculated with soybeans at 45 °C for 72 h. The sensory evaluation of the fermented soybeans was performed by well-trained panelists on five components that included two appearance components (color and stickiness), two aroma components (ammonia and soy sauce-like aroma), and texture, with the individual scores added together to provide a total score (Additional file 2: Table S5). According to the sensory evaluation, it was found that the soy sauce-like aroma of fermented soybeans with BJ3-2ΔrocF was prominent, whereas the ammonia was decreased (Additional file 2: Table S5). The degree of browning and viscosity were decreased in BJ3-2ΔrocF and BJ3-2ΔrocF + Arg (Fig. 5A, B) (Additional file 1: Fig. S6). Moreover, the pH of the fermented soybeans with BJ3-2ΔrocF was significantly lower than that of the fermented soybeans with BJ3-2 (Fig. 5C), indicating that knockout of rocF induced changes in the pH of the fermentation environment. In addition, the OD600 values of soybean slurries fermented with BJ3-2 and BJ3-2ΔrocF were 0.518 and 0.501, respectively (Fig. 5D), suggesting that the OD600 value of the soybean slurry fermented with BJ3-2ΔrocF was significantly reduced compared to that of BJ3-2.
Fermented soybeans by BJ3-2 and BJ3-2ΔrocF. A Fermented soybeans by BJ3-2 at 45 °C; B fermented soybeans by BJ3-2ΔrocF at 45 °C; C pH values of fermented soybeans by BJ3-2 and BJ3-2ΔrocF; D OD600 of fermented soybeans by BJ3-2 and BJ3-2ΔrocF. In (C) and (D), values represent mean ± standard deviation obtained from independent triplicates. Different letters indicate significance difference amongst the various treatments (P < 0.05)
rocF decreased the production of ammonia
Following the sensory evaluation, we found that the ammonia of fermented soybeans with BJ3-2ΔrocF was decreased. Therefore, we further detected the ammonia content of the fermented soybeans by a gas detector. The results showed that the ammonia content of fermented soybeans with BJ3-2ΔrocF was 137.7 ppm, while the value was 241.6 ppm in BJ3-2, which was decreased by 43% (Table 5). The results indicated that the rocF gene affects the products of ammonia.
rocF enhanced the contents of arginine, NH4 +, and TTMP in fermented soybeans
Indeed, arginine was converted into urea and ornithine through the urea cycle [26]. Therefore, the arginine content was measured by HPLC methodologies. The arginine content of fermented soybeans with BJ3-2ΔrocF was 0.590 mg/mL, which was dramatically increased compared to those fermented with BJ3-2 (Table 5). Subsequently, the content of NH4+ was detected by ion chromatography. A significant increase was obtained in fermented soybeans with BJ3-2ΔrocF (0.410 mg/kg) compared to those fermented with BJ3-2 (0.297 mg/kg) (Table 5). In addition, we also observed that the content of TTMP was increased by 8.6% in the fermented soybeans with BJ3-2ΔrocF (0.4617 mg/g) compared to those fermented with BJ3-2 (0.425 mg/g) (Table 6). Furthermore, a significantly increased content of TTMP in the BJ3-2ΔrocF treated with arginine were observed (Table 6), which indicated that rocF affects TTMP metabolic flow through the arginine pathway. However, the knockout of rocF resulted in slightly higher accumulations of the acetoin (Table 6). To sum up, these results indicated that the rocF gene affected the products of TTMP.
Discussion
B. subtilis is an aerobic, Gram-positive soil bacterium widely used in the food industry [6, 27, 28]. B. subtilis BJ3-2, used in this study, was isolated from fermented soybeans [29]. Fermented broth with BJ3-2 exhibited a prominent soy sauce-like aroma at 45 °C (Table 1). Furthermore, RNA-seq was performed on BJ3-2 (at 37 °C and 45 °C). The dominant DEGs were enriched in pyrimidine metabolism and arginine and proline metabolism (Additional file 2: Table S4). Most DEGs, including carA, pyrAa, pyrC, pyrDI, pyrDII, carB (BSU11240), pyrE, carB (BSU15520), pyrF, and pyrB, were involved in pyrimidine metabolism. The deficiency of each gene affects pyrimidine biosynthesis and ultimately leads to pyrimidine-deficient strains [30,31,32]. Moreover, eight DEGs (fadM, argF, argD, rocD, rocF, speD, rocG, and rocA) were enriched in arginine and proline metabolism (Table 4). Arginine metabolism is closely related to the products of TTMP, which contributes to the soy sauce-like aroma [33]. Among them, the expression of three genes (fadM, argF, and argD) was up-regulated at least 1 ~ fold (log2FC), and five genes (rocD, rocF, sped, rocG, and rocA) were down-regulated at least 3 ~ fold (log2FC) after incubation at 45 °C. The rocD was the most significantly down-regulated gene (4.32 ~ fold), while the knockdown of rocD resulted in arginine-deficient strains [25]. Interestingly, the rocF of BJ3-2 was down-regulated 4.16 ~ fold at 45 °C compared to 37 °C (Table 4), indicating its potential function in producing the soy sauce-like aroma.
Recently, studies have found that the deletion of key genes in the microorganisms may harm strain growth. For example, the knockout of the surfactin synthetase (srf) gene seems not to be favorable for strain growth in B. subtilis PB2-L [34]. However, in this study, the growth of the BJ3-2ΔrocF strain showed no significant changes in color and morphology, and growth curves compared to BJ3-2 (Fig. 3), suggesting the knockout of the rocF gene does not affect the normal growth of BJ3-2. However, the arginase activity was significantly inhibited in BJ3-2ΔrocF (Fig. 4), suggesting knockout of rocF only contributes to the inhibition of the arginase activity, but not change the growth characteristics of BJ3-2. Moreover, nonenzymatic browning is important in evaluating the Maillard reaction products (MRPs) [35, 36], which is affected by the reaction time, temperature, pH, solvent, and other conditions [37,38,39,40]. In the final stage of the Maillard reaction, the browning intensity is enhanced by increasing the initial pH value and other conditions [41]. In our study, the fermented soybeans with BJ3-2ΔrocF exhibited a lighter color and a slightly decreased pH (Fig. 5). These results are consistent with a number of previous studies on the Maillard reaction.
Zhao et al.reported a strong relationship between the amino acids and pyrazines [23]. In our study, the arginine content in fermented soybeans with BJ3-2ΔrocF was dramatically increased compared with BJ3-2 (Table 5), indicating the rocF gene affects arginine metabolism. In addition, we found that deletion of rocF gene changes the expression of arginine and proline metabolic pathway genes (Additional file 2: Table S4). Therefore, we speculated that rocF participates in the arginine and proline metabolic pathway and may convert arginine into urea and ornithine in the urea cycle (Fig. 6). Moreover, Ra**i et al.reported that the production of TTMP is closely associated with amino acid metabolism and glycolytic pathways [12]. The amino acid metabolic pathway mainly provides a nitrogen source (NH3 or NH4+), and the glycolytic pathway mainly provides the precursor compound (acetoin) for the production of TTMP [http://www.genome.jp/kegg/) pathway enrichment analysis was performed for each DEG [43]. The raw reads and processed RNA-seq data in this work have been deposited in the NCBI Gene Expression Omnibus (GEO) database under record number GSE166082.
Reverse transcription-quantitative real-time PCR (RT-qPCR)
To detect rocF gene expression, the total RNA extracted above was used to synthesize the first chain cDNA using the StarScript II First-strand cDNA Synthesis Mix with gDNA Remover Kit (GenStar, Bei**g) following the manufacturer’s recommendations. Then, the cDNA was diluted to 200 ng/µl, and RT-qPCR was performed using a CFX96 Touch PCR instrument (Bio-Rad, USA). The composition of the reaction mixture and conditions were the same as previously described in Zhang’s study [44]. Each RT-qPCR analysis experiment was performed three times, and the primers used in this study are listed in Table S1. 16S rRNA was used as a reference gene for expression analysis.
Homologous recombination knockout vector construction and transformation
The HLarm and HRarm correspond to the homologous left and right arms of rocF, whereas cm corresponds to the chloramphenicol gene. DEDP is a double-exchange detection primer (Additional file 2: Table S1) for assessing the success of the transformation.
The HLarm and HRarm of rocF were amplified from the B. subtilis BJ3-2 genome. The cm was amplified from the pHT01cas9-p43 vector. The primers used for amplification are listed in Table S1. HLarm, cm, and HRarm were double-digested with the restriction endonucleases Sac I and BamH I, BamH I and Xba I, and Xba I and Hind III, respectively. The digested fragments of HLarm, cm, and HRarm were sequentially connected to the pUC18 vector. Finally, the homologous recombination knockout vector pUC18-HLarm-cm-HRarm was constructed. The recombinant plasmid was then transformed to B. subtilis BJ3-2 according to the description in the literature [45, 46]. The transformants were verified by PCR using DEDP primers (Additional file 2: Table S1). The genomic DNA of the transformants was extracted using the Bacterial DNA Kit (OMEGA, USA) following the manufacturer’s recommendations and sequenced by the Sangon Biotech Co., Ltd. (Shanghai, China).
Enzyme determination assays
For the arginase determination assays, 0.1 g of BJ3-2 and BJ3-2ΔrocF bacteria were collected into a 1.5 mL centrifuge tube, sterile PBS (pH = 7.2) was added and broken by ultrasound in an ice bath and subjected to centrifugation (8,000 g for 5 min) to obtain the supernatant. The supernatant was transferred to a new tube used for the further assay. The arginase activity in BJ3-2 and BJ3-2ΔrocF was analyzed using the MEIMIAN reagent kit (Shanghai, China) and MULTISKAN GO (Thermo Fisher Scientific, Wilmington, DE, USA) according to the manufacturer’s instructions. A series of 2.5U/L, 5 U/L, 10 U/L, 20 U/L and 40 U/L concentrations were used to generate a standard curve. Three independent experiments were performed for each sample. The assays were done at least in triplicate.
Fermentation experiments
BJ3-2ΔrocF and BJ3-2 were inoculated in 5 mL of liquid LB medium and subsequently incubated at 37 °C with shaking at 180 rpm for 12 h. The soybeans were autoclaved at 121 °C for 20 min. The above bacterial suspensions (OD600 = 0.465) of BJ3-2ΔrocF and BJ3-2 were inoculated in autoclaved soybeans (1%, v/m) and fermented at 45 °C for 72 h.
Sensory evaluation
To assess the fermentation characteristics, the BJ3-2 strain was inoculated in a liquid LB medium and incubated at 37 °C and 45 °C for 72 h. The sensory analysis of the fermented broth was evaluated by 10 trained expert sensory panelists. The sensory score was a total of 3 indicators, including soy sauce-like aroma, Chi-flavour and ammonia (Additional file 2: Table S2). The sensory analysis of fermented soybeans by different strains was evaluated by 10 trained expert sensory panelists. The sensory score was a total of 5 indicators, including color, stickiness, ammonia, soy sauce-like aroma and texture (Additional file 2: Table S3).
pH measurement
The pH test was performed according to the Chinese national standard (GB5009.237–2016). Briefly, slurry was created from the fermented soybeans (10 g) using ultrapure water (50 mL), and the pH of the slurry was measured using a standard laboratory pH meter (pHS-3C). Experiments were performed in triplicate.
OD600 detection for the color of fermented soybeans
Fermented soybeans (20 g) were weighed and placed into a beaker with 100 mL of ultrapure water. The viscoid was cleaned, and the excess water of the soybeans was absorbed. Five grams of cleaned soybeans were placed in a mortar and ground into the homogenate with a pestle. Then, 50 mL of ultrapure water was added and mixed in the above homogenate. The mixture was well stirred and centrifuged at 8,000 g for 10 min. The absorbance of the supernatant fluid was measured at OD600. Experiments were conducted in three times.
Ammonia contents of fermented soybeans
The ammonia content of the fermented soybeans was detected by a gas detector (WOST Co., Ltd., Shenzhen, China) following the manufacturer’s recommendations.
Acetoin and TTMP contents of fermented soybeans
The concentrations of acetoin and TTMP were analyzed by HPLC. In brief, 10 g fermented soybean with BJ3-2, BJ3-2ΔrocF and BJ3-2ΔrocF + Arg (treated with 0.05% arginine) were dissolved in 30 mL 60% ethanol with 0.1 g CaCl2 and sonicated for 30 min at 25 °C (100 W, 20 kHz), and then centrifuged at 9000 rpm/min for 10 min at 4 °C. The supernatant was filtered by a 0.22 μm membrane before injection. The analysis was performed using Agilent LC1260 liquid chromatography (LC) system (California, USA), which was equipped with a flame ionization detector, a capillary column of Shim-pack GIST C18-AQ 5 μm (4.6 I.D. × 250 mm, SHIMADZU, Japan). Water with a trifluoroacetate concentration of 0.05% was mixed with methanol in a ratio of 7:3 (v/v) and used as the mobile phase at a flow rate of 0.7 mL/min. The column oven was kept at 30 °C for 5 min, then programmed to 220 °C with a stepwise increase of 30 °C /min and maintained at 220 °C for 3 min. The injection volume was 5 μL.
Arginine content of fermented broth
BJ3-2ΔrocF and BJ3-2 were inoculated in liquid LB medium (1% v/v) and fermented at 45 °C for 72 h. Subsequently, 2 mL of fermented broth was collected and centrifuged at 10,000 g for 2 min. Then, the supernatant was collected and stored at 4 °C. The arginine content was analyzed using a Waters 1525 high-performance liquid chromatography (HPLC) system equipped with an automatic injector and an ultraviolet detector (UV) at 210 nm (Waters Corp., Milford, USA), according to the description in the literature [47]. Briefly, all fermented samples were treated with potassium ferrocyanide (106 g/L) and zinc sulfate (300 g/L) and then centrifuged at 2,000 g for 8 min. Next, the supernatant was filtered through a 0.22 μm filter membrane prior to injection for HPLC analysis. Each injection volume was set to 10 µl. Chromatographic separation was achieved using a Waters Atlantis C18 column (5 μm, 4.6 × 150 mm) (Waters, USA) at 30 °C. The mobile phase of HPLC was 20 mmol/L NaH2PO4, and the flow rate was 1.0 mL/min. A calibration curve was established to quantify arginine. The experiment was performed in three biological replicates.
NH4 + contents of fermented soybeans
Fermented soybeans (20 mg) were ground into homogenate using a mortar and pestle and dissolved in 30 mL of methanesulfonic acid (MSA) (20 mM). Then, the above solution was extracted with ultrasonication for 20 min and repeated three times. All suspensions were collected and diluted to 100 mL. The mixture was filtered with a 0.45 µm filter membrane. The NH4+ content was determined with ion chromatography according to the description in the literature [48]. Briefly, the samples were detected with an LC2010PLUS (Shimadzu, Japan) equipped with an EGC eluent generator and a DS6 conductivity detector. The chromatographic system was an IonPac CS11-HC column (4.0 × 250 mm) with an IonPac CG12A (4.0 × 50 mm) pre-column. The injection volume was set to 20 µl. The flow rate was 1.0 mL/min, and the suppression current was 59 mA. Experiments were conducted in triplicate.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
Abbreviations
- TTMP:
-
Tetramethylpyrazine.
- SSF:
-
Solid-state fermentation
References
**ao ZJ, **e NZ, Liu PH, Hua DL, Xu P. Tetramethylpyrazine production from glucose by a newly isolated Bacillus mutant. Appl Microbiol Biotechnol. 2006. https://doi.org/10.1007/s00253-006-0491-6.
Müller R, Rappert S. Pyrazines: occurrence, formation and biodegradation. Appl Microbiol Biotechnol. 2010. https://doi.org/10.1007/s00253-009-2362-4.
Fan WL, Yan X, Zhang YH. Characterization of pyrazines in some Chinese liquors and their approximate concentrations. J Agric Food Chem. 2007;55(24):9956–62. https://doi.org/10.1021/jf071357q.
**ao ZB, Dai SP, Niu YW, Yu HY, Zhu JC, Tian HX, Gu YB. Discrimination of Chinese vinegars based on headspace Solid-Phase Microextraction-Gas chromatography mass spectrometry of volatile compounds and multivariate analysis. J Food Sci. 2011. https://doi.org/10.1111/j.1750-3841.
Meng W, Wang RM, **ao DG. Metabolic engineering of Bacillus subtilis to enhance the production of tetramethylpyrazine. Biotechnol Lett. 2015. https://doi.org/10.1007/s10529-015-1950-x.
Xu Y, Jiang YF, Li XT, Sun BG, Teng C, Yang R, **ong K, Fan GS, Wang WH. Systematic characterization of the metabolism of acetoin and its derivative ligustrazine in Bacillus subtilis under micro-Oxygen conditions. J Agric Food Chem. 2018. https://doi.org/10.1021/acs.jafc.8b00113.
Meng W, **ao DG, Wang RM. Enhanced production of tetramethylpyrazine in Bacillus licheniformis BL1 by bdhA disruption and 2,3-butanediol supplementation. World J Microbiol Biotechnol. 2016. https://doi.org/10.1007/s11274-015-1992-1.
Wang YF, Zhang XJ, Xu CJ, Zhang GX. Synthesis and biological evaluation of danshensu and tetramethylpyrazine conjugates as cardioprotective agents. Chem Pharm Bull (Tokyo). 2017. https://doi.org/10.1248/cpb.c16-00839.
Fadel HHM, Lotfy SN, Asker MMS, Mahmoud MG, Al-Okbi Y. Nutty-like flavor production by Corynbacterium glutamicum1220T from enzymatic soybean hydrolysate. Effect of encapsulation and storage on the nutty flavoring quality. J Adv Res. 2018. https://doi.org/10.1016/j.jare.2018.01.003.
Zhu BF, Xu Y, Fan WL, Wu Q. High-yield fermentative preparation of tetramethylpyrazine by Bacillus sp. using an endogenous precursor approach. J Ind Microbiol Biotechnol. 2010. https://doi.org/10.1007/s10295-009-0661-5.
Wang M, Qin HL, Leng J, Zafar A, Amjad MW, Raja MAG, Hussain MA, Bukhari SNA. Synthesis and biological evaluation of new tetramethylpyrazine based chalcone derivatives as potential anti-alzheimer agents. Chem Biol Drug Des. 2018. https://doi.org/10.1111/cbdd.13355.
Ra**i KS, Aparna P, Sasikala C, Ramana CV. Microbial metabolism of pyrazines. Crit Rev Microbiol. 2011. https://doi.org/10.3109/1040841X.2010.512267.
Wang J, Zhong QP, Yang YY, Li HR, Wang L, Tong YG, Fang X, Liao ZL. Comparison of bacterial diversity between two traditional starters and the round-koji-maker starter for traditional cantonese Chi-flavor liquor brewing. Front Microbiol. 2019. https://doi.org/10.3389/fmicb.2018.01053.
Kosuge T, Adachi T, Kamiya H. Isolation of tetramethylpyrazine from culture of Bacillus natto, and biosynthetic pathways of tetramethylpyrazine. Nature. 1962. https://doi.org/10.1038/1951103a0.
Besson I, Creuly C, Gros JB, Larroche C. Pyrazine production by Bacillus subtilis in solid-state fermentation on soybeans. Appl Microbiol Biotechnol. 1997. https://doi.org/10.1007/s002530050961.
Larroche C, Gros JB. Special transformation processes using fungal spores and immobilized cells. Adv Biochem Eng Biotechnol. 1997. https://doi.org/10.1007/BFb0102066.
Larroche C, Besson I, Gros JB. High pyrazine production by Bacillus subtilis in solid substrate fermentation on ground soybeans. Process Biochem. 1999. https://doi.org/10.1016/S0032-9592(98)00141-1.
Zhang JJ, Zhao XY, Zhang JX, Zhao C, Liu JJ, Tian YJ, Yang LP. Effect of deletion of 2,3-butanediol dehydrogenase gene (bdhA) on acetoin production of Bacillus subtilis. Prep Biochem Biotechnol. 2017. https://doi.org/10.1080/10826068.2017.1320293.
Cui DY, Wei YN, Lin LC, Chen SJ, Feng PP, **ao DG, Lin X, Zhang CY. Increasing yield of 2,3,5,6-Tetramethylpyrazine in Baijiu through Saccharomyces cerevisiae metabolic engineering. Front Microbiol. 2020. https://doi.org/10.3389/fmicb.2020.596306.
**ao Z, Hou X, Lyu X, ** L, Zhao JY. Accelerated green process of tetramethylpyrazine production from glucose and diammonium phosphate. Biotechnol Biofuels. 2014. https://doi.org/10.1186/1754-6834-7-106.
**ao ZJ, Lu JR. Generation of acetoin and its derivatives in foods. J Agric Food Chem. 2014. https://doi.org/10.1021/jf5013902.
Xu YQ, Chu HP, Gao C, Tao F, Zhou ZK, Li K, Li LX, Ma CQ, Xu P. Systematic metabolic engineering of Escherichia coli for high-yield production of fuel bio-chemical 2,3-butanediol. Metab Eng. 2014. https://doi.org/10.1016/j.ymben.
Zhao GZ, Feng YX, Hadiatullah H, Zheng FP, Yao YP. Chemical characteristics of three kinds of Japanese soy sauce based on electronic senses and GC-MS analyses. Front Microbiol. 2021. https://doi.org/10.3389/fmicb.2020.579808.
Wen AY, Qin LK, Zeng HY, Zhu Y. Comprehensive evaluation of physicochemical properties and antioxidant activity of B. subtilis-fermented polished adlay subjected to different drying methods. Food Sci Nutr. 2020. https://doi.org/10.1002/fsn3.1508.
Hampel A, Huber C, Geffers R, Spona-Friedl M, Eisenreich W, Bange FC. Mycobacterium tuberculosis is a natural ornithine aminotransferase (rocD) mutant and depends on Rv2323c for growth on arginine. PLoS ONE. 2015. https://doi.org/10.1371/journal.pone.0136914.
Lu CD. Pathways and regulation of bacterial arginine metabolism and perspectives for obtaining arginine overproducing strains. Appl Microbiol Biotechnol. 2006. https://doi.org/10.1007/s00253-005-0308-z.
Li WX, Zhou XX, Lu P. Bottlenecks in the expression and secretion of heterologous proteins in Bacillus subtilis. Res Microbiol. 2004. https://doi.org/10.1016/j.resmic.2004.05.002.
Yan Z, Zheng XW, Chen JY, Han JS, Han BZ. Effect of different Bacillus strains on the profile of organic acids in a liquid culture of Daqu. Asymmetric Michael Add Alanine Deriv. 2013. https://doi.org/10.1002/jib.58.
Jia DX, Wu YJ. Screening and Identification of Bacillus for lobster sauce fermentation. Food Sci. 2009. https://doi.org/10.3321/j.issn:1002-6630.2009.05.051.
Ralli P, Srivastava AC, O’Donovan G. Regulation of the pyrimidine biosynthetic pathway in a pyrD knockout mutant of Pseudomonas aeruginosa. J Basic Microbiol. 2010. https://doi.org/10.1002/jobm.200610248.
Butcher BG, Chakravarthy S, D’Amico K, Stoos KB, Filiatrault MJ. Disruption of the carA gene in Pseudomonas syringae results in reduced fitness and alters motility. BMC Microbiol. 2016. https://doi.org/10.1186/s12866-016-0819-z.
Yang HJ, Bogomolnaya L, McClelland M, Andrews-Polymenis H. De novo pyrimidine synthesis is necessary for intestinal colonization of Salmonella Typhimurium in chicks. PLoS ONE. 2017. https://doi.org/10.1371/journal.pone.0183751.
Shen YF. Type of flavor of Baijiu. Niangjiu. 2003. CNKI:SUN:NJZZ.0.2003–01–000.
Liu HX, Qu XX, Zhao SM, Lu ZX, Zhang C, Bie XM. Characterization of a Bacillus subtilis surfactin synthetase knockout and antimicrobial activity analysis. J Biotechnol. 2016. https://doi.org/10.1016/j.jbiotec.2016.08.018.
Jalbout AF, Shipar MAH, Trzaskowski B, Adamowicz. Formation of pyrazines in hydroxyacetaldehyde and glycine nonenzymatic browning Maillard reaction: a computational study. Food Chem. 2007. https://doi.org/10.1016/j.foodchem.2006.07.061.
Li ZP, Ma DX, He YY, Guo SQ, Liu FG, Liu XB. Simultaneous ultrasound and heat enhance functional properties of glycosylated lactoferrin. Molecules. 2020. https://doi.org/10.3390/molecules25235774.
Scaman C, Nakai S, Aminlari M. Effect of pH, temperature and sodium bisulfite or cysteine on the level of Maillard-based conjugation of lysozyme with dextran, galactomannan and mannan. Food Chem. 2006. https://doi.org/10.1016/j.foodchem.2005.08.003.
Benjakul S, Lertittikul W, Bauer F. Antioxidant activity of Maillard reaction products from a porcine plasma protein-sugar model system. Food Chem. 2005. https://doi.org/10.1016/j.foodchem.2004.10.019.
Chen SL, ** SY, Chen CS. Relative reactivities of glucose and galactose in browning and pyruvaldehyde formation in sugar/glycine model systems. Food Chem. 2005. https://doi.org/10.1016/j.foodchem.2004.09.005.
Ramírez-Jiménez A, García-Villanova B, Guerra-Hernández E. Effect of toasting time on the browning of sliced bread. J Sci Food Agric. 2001. https://doi.org/10.1002/jsfa.840.
Liu PL, Lu XM, Li NY, Zheng ZJ, Qiao XG. Characterization, variables, and antioxidant activity of the Maillard reaction in a Fructose-Histidine model system. Molecules. 2018. https://doi.org/10.3390/molecules24010056.
Kanamori T, Kanou N, Atomi H, Imanaka T. Enzymatic characterization of a prokaryotic urea carboxylase. J Bacteriol. 2004. https://doi.org/10.1128/JB.186.9.2532-2539.2004.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe. KEGG: integrating viruses and cellular organisms. Nucl Acids Res. 2021. https://doi.org/10.1093/nar/gkaa970.
Zhang LC, Zhu MK, Ren LJ, Li AZ, Chen GP, Hu ZL. The SlFSR gene controls fruit shelf-life in tomato. J Exp Bot. 2018. https://doi.org/10.1093/jxb/ery116.
Zhang XZ, Cui ZL, Hong Q, Li SP. High-level expression and secretion of methyl parathion hydrolase in Bacillus subtilis WB800. Appl Environ Microbiol. 2005. https://doi.org/10.1128/AEM.71.7.4101-4103.2005.
Phan TT, Nguyen HD, Schumann W. Novel plasmid-based expression vectors for intra-and extracellular production of recombinant proteins in Bacillus subtilis. Protein Expr Purif. 2006;46:189–95. https://doi.org/10.1016/j.pep.2005.07.005.
Xu W, Xu Q, Chen JH, Lu ZM, **a R, Li GQ, Xu ZH, Ma YH. Ligustrazine formation in Zhenjiang aromatic vinegar: changes during fermentation and storing process. J Sci Food Agric. 2011. https://doi.org/10.1002/jsfa.4356.
Favaro G, Pastore P, Saccani G, Cavalli S. Determination of biogenic amines in fresh and processed meat by ion chromatography and integrated pulsed amperometric detection on Au electrode. Food Chem. 2007. https://doi.org/10.1016/j.foodchem.2007.04.071.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant No. 32060585) and the Application and Transformation Project of Scientific and Technological Achievements in Guizhou Province ([2019]4224).
Funding
This study was financially supported by Natural Science Foundation of China (NSFC).
Author information
Authors and Affiliations
Contributions
LZL and WYJ conceptualized and designed the study, drafted the initial manuscript, and reviewed and revised the manuscripted. ZLC, TSQ, JJ, GX and ZJ designed the data collection instruments, collected data, carried out and initial analyses, and reviewed and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
The authors provide consent for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1. Figure S1
. Data quality control; Figure S2. Expression density distribution; Figure S3. RT-qPCR of rocF; Figure S4. Construction of the homologous recombination knockout vector; Figure S5. Sequencing of BJ3-2ΔrocF; Figure S6. Fermented soybeans with BJ3-2, BJ3-2ΔrocF and BJ3-2ΔrocF+Arg at 45 °C
Additional file2. Table S1.
Primers used in the study; Table S2. Scoring standard for sensory evaluation of BJ3-2 at 37 °C and 45 °C ;Table S3. Scoring standard for sensory evaluation of fermented soybeans by different strains. Table S4. The analysis of KEGG pathway enrichment; Table S5. Sensory evaluation of fermented soybeans by different strains
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Liu, Z., Wu, Y., Zhang, L. et al. rocF affects the production of tetramethylpyrazine in fermented soybeans with Bacillus subtilis BJ3-2. BMC Biotechnol 22, 18 (2022). https://doi.org/10.1186/s12896-022-00748-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12896-022-00748-4