Abstract
Background
Nitraria sibirica Pall. is a halophytic shrub with strong environmental adaptability that can survive in extremely saline-alkali and drought-impacted environments. Gene expression analysis aids in the exploration of the molecular mechanisms of plant responses to abiotic stresses. RT–qPCR is the most common technique for studying gene expression. Stable reference genes are a prerequisite for obtaining accurate target gene expression results in RT–qPCR analysis.
Results
In this study, a total of 10 candidate reference genes were selected from the transcriptome of N. sibirica, and their expression stability in leaves and roots under different treatment conditions (salt, alkali, drought, cold, heat and ABA) was evaluated with the geNorm, NormFinder, BestKeeper, comparative ΔCt and RefFinder programs. The results showed that the expression stability of the candidate reference genes was dependent on the tissue and experimental conditions tested. ACT7 combined with R3H, GAPDH, TUB or His were the most stable reference genes in the salt- or alkali-treated leaves, salt-treated roots and drought-treated roots, respectively; R3H and GAPDH were the most suitable combination for drought-treated leaves, heat-treated root samples and ABA-treated leaves; DIM1 and His maintained stable expression in roots under alkali stress; and TUB combined with R3H was stable in ABA-treated roots. TBCB and GAPDH exhibited stable expression in heat-treated leaves; TBCB, R3H, and ERF3A were stable in cold-treated leaves; and the three most stable reference genes for cold-treated roots were TBCB, ACT11 and DIM1. The reliability of the selected reference genes was further confirmed by evaluating the expression patterns of the NsP5CS gene under the six treatment conditions.
Conclusion
This study provides a theoretical reference for N. sibirica gene expression standardization and quantification under various abiotic stress conditions and will help to reveal the molecular mechanisms that confer stress tolerance to N. sibirica.
Similar content being viewed by others
Background
Nitraria sibirica Pall. is a typical salt-diluting halophyte shrub that can adapt to diverse adverse stresses, such as high salinity, alkali, drought and extreme temperatures. Therefore, N. sibirica is often used as a pioneer tree for windbreaks, sand fixation and the improvement of heavily saline-alkali soils [1, 2]. As a sessile plant that grows in extreme environments, N. sibirica has had to develop a series of complex mechanisms to resist or adapt to adverse environmental conditions. In addition, N. sibirica can also serve as an economic plant in saline–alkali areas due to its medicinal value and edible fruit [9]. However, the accuracy and reliability of RT–qPCR results are affected by many factors, such as the quality of the RNA, the efficiency of cDNA synthesis, and the efficiency of amplification [10, 11]. To obtain accurate gene expression results, the data generated by RT–qPCR should be normalized with the use of appropriate reference genes [7, 11]. Housekee** genes that maintain the cytoskeleton or participate in basic biochemical metabolism, such as actin (ACT), β-tubulin (TUB), elongation factor 1-α (EF-1α) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), are usually selected as internal reference genes [12,13,14]. The expression levels of reference genes should ideally be consistent in different tissues and under different environmental conditions [15, 16]. However, numerous studies have found that the stability of reference gene expression is different in different species, tissues and experimental treatments [17, 18]. For example, ACT was a stable reference gene in Platycladus orientalis [19] but unstable in teak (Tectona grandis L.f.) [20]. Ubiquitin conjugating enzyme (UBC) and GAPDH were the most stable reference genes in the flowers of Iris germanica L. ‘00246’ and ‘Elizabeth’, while the most stable reference genes in the flowers of Iris germanica L ‘2010200’ were TUB and UBC [21]. In Glehnia littoralis, expressed protein 1 (EXP1) and serine/threonine-protein phosphatase PP2A (PP2A) were the most stable reference genes under salt stress, while cyclophilin 2 (CYP2) and α-TUB were the most stable genes under MeJA treatment [22]. In addition, it has been reported that some newly characterized genes may be more stable than traditional housekee** genes. Under salt and drought stress, the expression of the new reference genes RG1, RG3 and RG5 in poplar was more stable than that of 18S ribosomal RNA (18S rRNA), Actin and ubiquitin (UBQ) [27], comparative ΔCt [28] and RefFinder [29]) were applied to evaluate their expression stability for normalization. In addition, the expression of Δ1-pyrroline-5-carboxylate synthetase (NsP5CS) was analysed to demonstrate the effectiveness of the selected reference genes. Our work will facilitate further studies of gene expression in N. sibirica and will accelerate the understanding of the molecular mechanisms underlying stress-induced responses in N. sibirica.
Results
Primer specificity and amplification efficiency test of candidate reference genes
A total of 10 genes were selected from transcriptome data and used as candidate reference genes. qPCR primers were designed based on these sequences. All primers showed a signal and clear band of the expected size, and there was no primer-dimer formation observed on gel electrophoresis (Fig. S1), indicating the specificity of the primer pairs. The single peaks presented in the melting curve assays of each gene further verified the specificity of the primer pairs (Fig. S2). The amplification efficiency of the 10 candidate reference genes ranged from 89.67% (His) to 113.81% (ACT11), and the correlation coefficients (R2) varied from 0.984 to 0.999 (Table 1). These results indicated that the primers of the 10 genes met the standards for qPCR and could be used in subsequent experiments. The details of the candidate reference genes, primer sequences, amplification lengths and efficiencies, and correlation coefficients are shown in Table 1.
Expression levels of candidate reference genes in leaves and roots
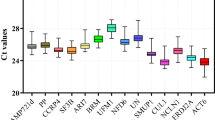
The quantification cycle (Cq) value reflects the mRNA transcript level. Reference genes with higher Cq values are considered to have lower expression abundance. Based on RT–qPCR, we determined the expression levels of 10 candidate reference genes in root and leaf tissues under salt, alkali, drought, cold, heat and ABA treatments. As shown in Fig. 1, the Cq values of the 10 candidate reference genes varied from 17.56 (EF-1α) to 29.95 (ERF3A), indicating a wide range of expression abundance. EF-1α and ERF3A were the most [mean Cq of 21.02 ± 2.46 (standard deviation, SD)] and least (mean Cq of 27.25 ± 1.66) expressed genes. In addition, it was observed that TBCB, R3H and DIM1 had relatively small SD values of 0.87, 0.99 and 1.01, respectively, whereas EF-1α and ERF3A showed relatively high variation, with SD values of 2.46 and 1.66, respectively (Fig. 1, Table S1). Further analysis of the distribution of Cq values showed that the Cq values of the 10 candidate reference genes varied differently in different tissues and treatments. For example, ACT11 and TUB had Cq values of 23.06 ± 0.93 and 23.48 ± 1.06 in leaves, respectively, but their Cq values in roots were 25.22 ± 0.90 and 24.72 ± 1.61, respectively (Fig. S3). The Cq value of EF-1α was 19.66 ± 1.13 under salt treatment, but the Cq value under heat treatment was 22.80 ± 3.14 (Fig. S4). Therefore, screening suitable reference genes for specific experimental treatments and tissues is needed.
Expression stability of candidate reference genes
Four specialized analytical tools, geNorm, NormFinder, BestKeeper and the comparative ΔCt method, were used to examine the expression stability of the 10 candidate reference genes. Subsequently, the RefFinder tool was employed to evaluate the expression stability of all these candidate reference genes and select the most suitable genes.
geNorm analysis
geNorm evaluated the stability of all 10 candidate reference genes using the M value (reference expression stability measure). The default threshold of M is 1.5. A lower M value indicates more stable gene expression [26]. As illustrated in Fig. 2, the M values of all 10 candidate reference genes in every treatment were lower than the default limit of 1.5, and all the candidate reference genes had different levels of stability under different treatments. ACT7 and R3H, which had the same M values, were the most stable genes for salt-treated leaves (SL), drought-treated leaves (DL) and drought-treated roots (DR); TUB showed good stability for salt-treated roots (SR), alkali-treated roots (AR), heat-treated roots (HR) and abscisic acid-treated roots (ABR); and GAPDH was one of the most stable genes in SR, alkali-treated leaves (AL), heat-treated leaves (HL) and abscisic acid-treated leaves (ABL). EF-1α and His were determined to be the least stable reference genes across treatments. geNorm software also provides information on the optimum number of reference genes to be used in an experiment based on the pairwise variation between ranked genes (Vn/n + 1), and a cut-off of 0.15 (V value) is usually applied to determine whether an additional reference gene needs to be added [26]. If Vn/n + 1 > 0.15, then n + 1 reference genes need to be used; otherwise, only n reference genes are needed. As shown in Fig. 3, the V2/3 values for the SL (0.054), SR (0.071), AL (0.068), AR (0.081), DL (0.106), DR (0.100), ABL (0.142), ABR (0.097), HL (0.123) and HR (0.145) samples were lower than 0.15 (Fig. 3), indicating that two reference genes would be sufficient for the accurate normalization of these treatment samples. The V2/3 values for cold-treated leaves (CL) and cold-treated roots (CR) exceeded 0.15, while the V3/4 values were 0.110 and 0.140, respectively, indicating that three reference genes were needed.
Expression stability values (M) of 10 candidate reference genes validated via the geNorm program. The least stable genes with higher M-values are on the left, and the most stable genes with lower M-values are on the right. SL and SR: Salt-treated leaves and roots, respectively; AL and AR: alkali-treated leaves and roots, respectively; DL and DR: drought-treated leaves and roots, respectively; CL and CR: cold-treated leaves and roots, respectively; HL and HR: heat-treated leaves and roots, respectively; ABL and ABR: abscisic acid-treated leaves and roots, respectively
NormFinder analysis
NormFinder software directly evaluates the stability of reference genes according to the variance within and between groups, with lower values indicating higher stability [10]. The stability values of the candidate reference genes in each treatment are shown in Fig. 4 and Table S2. The stability rankings of the 10 reference genes under different experimental conditions were not completely consistent. For example, ACT7 was the most stable reference gene in the SL samples, while TUB was ranked first in the SR and ABR samples, and GAPDH was the top ranked gene for the AL, DL and HR samples. The stability ranking of the 10 candidate reference genes generated with NormFinder was slightly different from that produced by geNorm in most of the samples. For example, His and ACT11 were the top ranked reference genes for the DR and CR samples in the NormFinder analysis, while they ranked third and fourth, respectively, in the geNorm analysis.
BestKeeper analysis
BestKeeper evaluates the stability of gene expression by calculating the standard deviation (SD) and coefficient of variation (CV) of the Cq values. A smaller SD and CV indicate better stability of gene expression. If SD > 1, the gene was considered unstable [27]. The analysis results are listed in Fig. 4 and Table S3. GAPDH was the most stable gene in the AL samples. His and TUB were ranked as the most stably expressed genes in the SR and DR samples, respectively, but exhibited the lowest stability in the HL and SL samples, respectively. DIM1 was the most stable reference gene in the SL and HL samples, and ACT11 was the most stable reference gene in the HR and ABR samples. For the AR, DL, CL and CR samples, the most stable gene was TBCB. EF-1α was ranked as the least stably expressed gene in most of the samples, including the AL, AR, DL, DR, CL, HL, HR and ABL samples. Overall, the stability ranking of the 10 candidate reference genes generated with BestKeeper was different from that of geNorm and NormFinder.
Comparative ΔCt analysis
This method evaluated gene expression stability by calculating the mean standard deviation (SD) value of each gene. Here, the smaller the value is, the higher the stability [28]. In SL, AL, AR, DR, HL, and ABR samples, ACT7 was one of the two best reference genes for gene normalization. For ABL, DL, CL, and HR samples, one of the two most stable reference genes was R3H. In addition, the comparative ΔCt analysis also showed that EF-1α was the least stably expressed gene in multiple samples, such as AL, DR, CL and HR samples (Fig. 4 and Table S4).
RefFinder analysis
To reduce the influence of the limitations of a single algorithm, comprehensive stability rankings of the candidate reference genes were determined with the RefFinder program (https://www.heartcure.com.au/reffinder/). The ranking of genes was computed as the geometric mean, and a lower geometric, a higher stability []. As shown in Fig. 4 and Table S5, ACT7 combined with R3H, TUB, GAPDH or His were the two most stable reference genes in the SL, SR, AL, and DR samples; R3H and GAPDH were suggested to be the most suitable combination for DL, ABL, and HR samples; for AR samples, the two most stable reference genes were DIM1 and His; and TUB and R3H were the most stable reference genes for ABR samples. TBCB and GAPDH were the most stable reference genes for HL samples; TBCB, R3H, and ERF3A were found to be the three most stable reference genes in CL samples; and the three most stable reference genes for CR samples were TBCB, ACT11 and DIM1. EF-1α was the least stable reference gene in most samples.
Reference gene validation
To verify the results generated through the analyses described above, the expression pattern of NsP5CS was examined in SL, DR, AL, CR, HL and ABR samples. P5CS is a rate-limiting enzyme in proline biosynthesis and plays an important role in controlling plant stress tolerance [30, 31]. The two most stable reference genes (alone and in combination) and the two most unstable reference genes based on the comprehensive ranking results for each sample set were used in the validation test. As shown in Fig. 5, the relative expression levels of NsP5CS normalized with the two most stable reference genes in combination showed significant changes in SL, DR, CR and ABR samples and different trends among different sample sets. In the SL and CR samples, NsP5CS was continuously induced, and its relative expression reached the highest level at 48 h, with values of 9.08 and 6.45, respectively. In DR samples, NsP5CS was rapidly induced and reached a maximum expression level at 3 h and then fluctuated at a lower level with time. NsP5CS was also rapidly induced and reached a maximum value at 3 h and remained at a higher value at 6 h for ABR samples. The expression patterns of NsP5CS normalized with the two most stable reference genes alone or in combination exhibited consistency in SL, DR, CR and ABR samples. However, the expression patterns of NsP5CS were significantly different when unstable reference genes were used. In the AL and HL samples, the expression level of NsP5CS varied less, but there were also similar expression patterns of NsP5CS normalized by the two most stable reference genes (alone or in combination) and were significantly different by the two unstable reference genes.
Relative expression of NsP5CS at 0, 3, 6, 12, 24, and 48 h following stress treatment using the selected reference genes for normalization. The validated reference gene(s) used as normalization factors were the two most stable reference genes (alone or in combination) and the two most unstable reference genes in different treatment groups. Error bars show the standard error calculated from three biological replicates
Discussion
RT–qPCR is one of the primary methods used to detect gene expression and thereby help to reveal the response mechanism of plants under different stresses [24] and poplar [13]. This result indicates that the expression level of the same gene may not be constant across multiple species [15]. In addition, GAPDH was the best reference gene in the AL, DL, ABL, HL and HR samples of N. sibirica, while it exhibited poor stability in SL and AR. The same reference gene in the same species may respond differently to different stresses, as in the case in Kentucky bluegrass [38] and A. stolonifera [9]. In summary, it is usually necessary to select reference genes specifically according to species, tissues, and treatments.
Numerous studies have reported significant variations in the expression levels of target genes when unstable reference genes were used as internal controls [57] was used to design the candidate reference gene primers. The parameters were set as follows: melting temperature (Tm), 55–65℃; GC content, 40–60%; primer length, 17–26 bp; and amplification product size, 150–300 bp [21, 58]. All primers (Table 1) were synthesized by RuiBiotech company (Bei**g, China). All primers were verified by electrophoresis on a 1.5% agarose gel.
RT–qPCR conditions and amplification efficiency test
RT–qPCR was performed in 96-well plates by using a LightCycler® 480II Real-Time PCR System (Roche Molecular Systems, Germany). cDNA was amplified by using UItraSYBR Mixture (CoWin Biosciences, Bei**g, China). Each reaction mixture contained 10 µl of 2×UItraSYBR Mixture, 0.8 µl of cDNA, 0.4 µl of each primer (10 µM) and 8.4 µl of ddH2O. The amplification conditions were as follows: an initial step of 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s, 55 °C for 30 s and 72 °C for 32 s. The final melting curve was produced by shifting the amplification temperature from 65 to 95 °C. RT–qPCR analysis of each sample was performed in triplicate, and template-free controls were included in parallel. The standard curve was constructed with a tenfold dilution (10, 100, 1000, 10,000) of a cDNA mixture comprising equal volumes of cDNA from all samples, and the amplification efficiency (E) and correlation coefficient (R2) values of the primers were calculated using the standard curve. The E value of each primer pair was calculated by the curve slope using E = 10(−1/slope) [59].
Stability analysis of candidate reference genes
To assess the stability of candidate reference genes, we first analysed the resulting RT–qPCR data using four software programs: geNorm, NormFinder, BestKeeper and comparative ΔCt method. Then, RefFinder (https://www.heartcure.com.au/reffinder/) was used to generate a comprehensive ranking of the candidate reference genes according to data obtained by geNorm, NormFinder, BestKeeper and comparative ΔCt [42]. For geNorm and NormFinder analysis, the raw quantification cycle values (Cq) need to be converted into relative quantities, and for BestKeeper and comparative ΔCt algorithms, raw Cq values could meet the requirements.
Validation of reference gene stability
To validate the reliability of the selected reference genes, the relative expression level of NsP5CS was analysed in SL, AL, DR, CR, HL and ABR samples. The primer pair of NsP5CS (Table 1) was designed with Primer Premier 5.0 software. The RT‒qPCR conditions were set up the same as the RT‒qPCR conditions described above. The expression of NsP5CS was calculated using (Etarget) ΔCqtarget(control−sample)/(Eref) ΔCqref(control−sample) [60] with the two worst and two best reference genes (alone and in combination) obtained by the comprehensive assessment used as references.
Availability of data and materials
The main data supporting the results of this article are included within the article (and its additional files).
References
Zhang G. Effects of iso-osmotic salt and water stresses on growth and ionic absorption and distribution in Nitraria sibirica seedlings. Agric Res Arid Areas. 2013;31(4):114–18.
Li H, Tang X, Zhu J, Yang X, Zhang H. De novo transcriptome characterization, gene expression profiling and ionic responses of Nitraria sibirica Pall. Under salt stress. Forests. 2017;8(6):211.
Song Q, **a X, Ji C, Chen D, Lu Y. Optimized flash extraction and UPLC-MS analysis on antioxidant compositions of Nitraria sibirica fruit. J Pharm Biomed Anal. 2019;172:379–87.
Sa R, chen G. Effect of exogenous spermidine on antioxidant enzyme system in leaves of Nitraria sibirica Pall. Seedlings under salt stress. Acta Bot Boreali-Occidentalia Sin. 2013;33(02):352–56.
Chen T, Li H, Wu h, Liu z, Wu X, Yang S, et al. Comparison on osmotica accumulation of different salt-tolerant plants under salt stress. For Res. 2015;28(06):826–32.
Tang X, Zhang H, Shabala S, Li H, Yang X, Zhang H. Tissue tolerance mechanisms conferring salinity tolerance in a halophytic perennial species Nitraria sibirica Pall. Tree Physiol. 2021;41(7):1264–77.
Derveaux S, Vandesompele J, Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–30.
Kanakachari M, Solanke AU, Prabhakaran N, Ahmad I, Dhandapani G, Jayabalan N, et al. Evaluation of suitable reference genes for normalization of qPCR gene expression studies in Brinjal (Solanum melongena L.) during Fruit Developmental Stages. Appl Biochem Biotechnol. 2016;178(3):433–50.
Chen Y, Hu B, Tan Z, Liu J, Yang Z, Li Z, et al. Selection of reference genes for quantitative real-time PCR normalization in cree** bentgrass involved in four abiotic stresses. Plant Cell Rep. 2015;34(10):1825–34.
Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50.
Delporte M, Legrand G, Hilbert JL, Gagneul D. Selection and validation of reference genes for quantitative real-time PCR analysis of gene expression in Cichorium intybus. Front Plant Sci. 2015;6:651.
Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6(6):609–18.
Wang B, Duan H, Chong P, Su S, Shan L, Yi D, et al. Systematic selection and validation of suitable reference genes for quantitative real-time PCR normalization studies of gene expression in Nitraria tangutorum. Sci Rep. 2020;10(1):15891.
Shukla P, Reddy RA, Ponnuvel KM, Rohela GK, Shabnam AA, Ghosh MK, et al. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses. Mol Biol Rep. 2019;46(2):1809–17.
Sgamma T, Pape J, Massiah A, Jackson S. Selection of reference genes for diurnal and developmental time-course real-time PCR expression analyses in lettuce. Plant Methods. 2016;12:21.
He Y, Zhong Y, Bao Z, Wang W, Xu X, Gai Y, et al. Evaluation of Angelica decursiva reference genes under various stimuli for RT-qPCR data normalization. Sci Rep. 2021;11(1):18993.
Hu X, Zhang L, Nan S, Miao X, Yang P, Duan G, et al. Selection and validation of reference genes for quantitative real-time PCR in Artemisia sphaerocephala based on transcriptome sequence data. Gene. 2018;657:39–49.
Yang C-L, Yuan X-Y, Zhang J, Sun W-H, Liu Z-J, Zou S-Q. Comprehensive transcriptome analysis of reference genes for fruit development of Euscaphis konishii. PeerJ. 2020;8:e8474.
Chang E, Shi S, Liu J, Cheng T, Xue L, Yang X, et al. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS ONE. 2012;7(3):e33278.
Galeano E, Vasconcelos TS, Ramiro DA, De Martin Vde F, Carrer H. Identification and validation of quantitative real-time reverse transcription PCR reference genes for gene expression analysis in teak (Tectona grandis L.f.). BMC Res Notes. 2014;7:464.
Wang Y, Zhang Y, Liu Q, Tong H, Zhang T, Gu C, et al. Selection and validation of appropriate reference genes for RT-qPCR analysis of flowering stages and different genotypes of Iris germanica L. Sci Rep. 2021;11(1):9901.
Li L, Li N, Fang H, Qi X, Zhou Y. Selection and validation of reference genes for normalisation of gene expression in Glehnia littoralis. Sci Rep. 2020;10(1):7374.
Chu W, Wang Y, Zhu D, Chen Z, Yan H, **ang Y. Selection of novel reference genes in Poplar under salt and drought stresses. Sci Silvae Sin. 2017;53(10):70–79.
Liu Q, Qi X, Yan H, Huang L, Nie G, Zhang X. Reference gene selection for quantitative real-time reverse-transcriptase PCR in Annual Ryegrass (Lolium multiflorum) subjected to various abiotic stresses. Molecules. 2018;23(1):172.
Bakir Y, Eldem V, Zararsiz G, Unver T. Global transcriptome analysis reveals differences in gene expression patterns between nonhyperhydric and hyperhydric peach leaves. Plant Genome. 2016;9(2):0080.
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):467–72.
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekee** genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–15.
Silver N, Best S, Jiang J, Thein SL. Selection of housekee** genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33.
**e F, **ao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012. doi:https://doi.org/10.1007/s11103-012-9885-2.
Guan C, Ji J, Guan W, Feng Y, Li X, ** C, et al. A Lycium chinense-derived P5CS-like gene is regulated by water deficit-induced endogenous abscisic acid and overexpression of this gene enhances tolerance to water deficit stress in Arabidopsis. Mol Breeding. 2014;34(3):1109–24.
Zheng L, Dang Z, Li H, Zhang H, Wu S, Wang Y. Isolation and characterization of a ∆1-pyrroline-5-carboxylate synthetase (NtP5CS) from Nitraria tangutorum Bobr. And functional comparison with its Arabidopsis homologue. Mol Biol Rep. 2014;41(1):563–72.
**ao X, Ma J, Wang J, Wu X, Li P, Yao Y. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front Plant Sci. 2014;5:788.
Niu X, Chen M, Huang X, Chen H, Tao A, Xu J, et al. Reference gene selection for qRT-PCR normalization analysis in kenaf (Hibiscus cannabinus L.) under abiotic stress and hormonal stimuli. Front Plant Sci. 2017;8:771.
Bustin S. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29(1):23–39.
Taylor S, Wakem M, Dijkman G, Alsarraj M, Nguyen M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods. 2010;50(4):S1–S5.
Abbas A, Yu H, Li X, Cui H, Chen J, Huang P. Selection and validation of reference genes for RT-qPCR analysis in Aegilops tauschii (Coss.) Under different abiotic stresses. Int J Mol Sci. 2021;22(20):10017.
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399(2):257–61.
Niu K, Shi Y, Ma H. Selection of candidate reference genes for gene expression analysis in Kentucky Bluegrass (Poa pratensis L.) under abiotic stress. Front Plant Sci. 2017;8:193.
Yang Y, Zhao L, Yang G, ZHANG Y, Fu P, Hu J, et al. Selection and validation of reference genes for leaf color phenotype in ‘Maiyuan**qiu’, a Catalpa fargesii variety, by qRT-PCR. For Res. 2022;35(1):123–31.
Zhao J, Zhou M, Meng Y. Identification and validation of reference genes for RT-qPCR analysis in Switchgrass under heavy metal stresses. Genes (Basel). 2020;11(5):502.
Feng K, Liu J, **ng G, Sun S, Li S, Duan A, et al. Selection of appropriate reference genes for RT-qPCR analysis under abiotic stress and hormone treatment in celery. PeerJ. 2019;7:e7925.
Su W, Yuan Y, Zhang L, Jiang Y, Gan X, Bai Y, et al. Selection of the optimal reference genes for expression analyses in different materials of Eriobotrya japonica. Plant Methods. 2019;15:7.
Dudziak K, Sozoniuk M, Szczerba H, Kuzdralinski A, Kowalczyk K, Borner A, et al. Identification of stable reference genes for qPCR studies in common wheat (Triticum aestivum L.) seedlings under short-term drought stress. Plant Methods. 2020;16:58.
Niaz Z, Sui Z, Riaz S, Liu Y, Phycology JJJoA. Identification of valid reference genes for the normalization of RT-qPCR gene expression data in Alexandrium catenella under different nutritional conditions. J Appl Phycol. 2019;31(1):1819–33.
Wang M, Ren T, Marowa P, Du H, Xu Z. Identification and selection of reference genes for gene expression analysis by quantitative real-time PCR in Suaeda glauca’s response to salinity. Sci Rep. 2021;11(1):8569.
Tang F, Chu L, Shu W, He X, Wang L, Lu M. Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in Poplar. Plant Methods. 2019;15:35.
Hou S, Zhao T, Yang D, Li Q, Liang L, Wang G, et al. Selection and validation of reference genes for quantitative RT-PCR analysis in Corylus heterophylla Fisch. X Corylus avellana L. Plants (Basel). 2021;10(1):159.
Li H, Tang X, Yang X, Zhang H. Comprehensive transcriptome and metabolome profiling reveal metabolic mechanisms of Nitraria sibirica Pall. To salt stress. Sci Rep. 2021;11(1):12878.
Yang Z, Zhang R, Zhou Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes. 2021;12(5):732.
Wei M, Chen Y, Zhang M, Yang J, Lu H, Zhang X, et al. Selection and validation of reference genes for the qRT-PCR assays of Populus ussuriensis gene expression under abiotic stresses and related ABA treatment. Forests. 2020;11(4):476.
Liang X, Zhang L, Natarajan SK, Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal. 2013;19(9):998–1011.
Su M, Li XF, Ma XY, Peng XJ, Zhao AG, Cheng LQ, et al. Cloning two P5CS genes from bioenergy sorghum and their expression profiles under abiotic stresses and MeJA treatment. Plant Sci. 2011;181(6):652–9.
Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, et al. Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995;7(5):751–60.
Song T, Sun N, Dong L, Cai H. Enhanced alkali tolerance of rhizobia-inoculated alfalfa correlates with altered proteins and metabolic processes as well as decreased oxidative damage. Plant Physiol Biochem. 2021;159:301–11.
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22.
Hao M, Wei C, Ren A, Li P. Seedling raising technique of Nitraria plants in Yellow River Delta. Sci Technol. 2015;25(18):92.
Liu Y, Zhang Y, Liu F, Liu T, Chen J, Fu G, et al. Establishment of reference (housekee**) genes via quantitative real-time PCR for investigation of the genomic basis of abiotic stress resistance in Psammochloa villosa (Poaceae). J Plant Physiol. 2022;268:153575.
Zhang Y, Zhu L, Xue J, Yang J, Hu H, Cui J, et al. Selection and verification of appropriate reference genes for expression normalization in Cryptomeria fortunei under abiotic stress and hormone treatments. Genes (Basel). 2021;12(6):791.
Meuer S, Wittwer C, Nakagawara K. Rapid cycle real-time PCR: methods and applications. Heidelberg: Springer Press; 2001. doi:21–34.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(900):2002–07.
Acknowledgements
We thank State Key Laboratory of Tree Genetics and Breeding, Research Institute of Forestry, Chinese Academy of Forestry for providing the experimental platform and those who contributed to this article.
Funding
This research was supported by the National Natural Science Foundation of China (31870663), the Science and Technology Program of Tangshan (20150211C) and the Fundamental Research Funds of CAF (project no. CAFYBB2020MA007). The funders played no role in designing the study, collecting and analysing the data, writing the manuscript, or deciding the publish.
Author information
Authors and Affiliations
Contributions
AH, JZ, HLZ, XW and HZ conceived and designed the experiments. AH, HLZ and JL performed the experiments. XY help to collect experimental materials and provide exprerimental instruments. HLZ, JW and HW critically revised the manuscript. The author(s) read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Figure S1, Figure S2, Figure S3 and Figure S4.
Additional file 2:
Table S1, Table S2, Table S3, Table S4 and Table S5.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hu, A., Yang, X., Zhu, J. et al. Selection and validation of appropriate reference genes for RT–qPCR analysis of Nitraria sibirica under various abiotic stresses. BMC Plant Biol 22, 592 (2022). https://doi.org/10.1186/s12870-022-03988-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03988-w