Abstract
Background
Cotton fiber is an important natural resource for textile industry and an excellent model for cell biology study. Application of glabrous mutant cotton and high-throughput sequencing facilitates the identification of key genes and pathways for fiber development and cell differentiation and elongation. LncRNA is a type of ncRNA with more than 200 nt in length and functions in the ways of chromatin modification, transcriptional and post-transcriptional modification, and so on. However, the detailed lncRNA and associated mechanisms for fiber initiation are still unclear in cotton.
Results
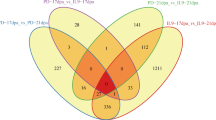
In this study, we used a novel glabrous mutant ZM24fl, which is endowed with higher somatic embryogenesis, and functions as an ideal receptor for cotton genetic transformation. Combined with the high-throughput sequencing, fatty acid pathway and some transcription factors such as MYB, ERF and bHLH families were identified the important roles in fiber initiation; furthermore, 3,288 lncRNAs were identified, and some differentially expressed lncRNAs were also analyzed. From the comparisons of ZM24_0 DPA vs ZM24_-2 DPA and fl_0 DPA vs ZM24_0 DPA, one common lncRNA MSTRG 2723.1 was found that function upstream of fatty acid metabolism, MBY25-mediating pathway, and pectin metabolism to regulate fiber initiation. In addition, other lncRNAs MSTRG 3390.1, MSTRG 48719.1, and MSTRG 31176.1 were also showed potential important roles in fiber development; and the co-expression analysis between lncRNAs and targets showed the distinct models of different lncRNAs and complicated interaction between lncRNAs in fiber development of cotton.
Conclusions
From the above results, a key lncRNA MSTRG 2723.1 was identified that might mediate some key genes transcription of fatty acid metabolism, MYB25-mediating pathway, and pectin metabolism to regulate fiber initiation of ZM24 cultivar. Co-expression analysis implied that some other important lncRNAs (e.g., MSTRG 3390.1, MSTRG 48719.1, and MSTRG 31176.1) were also showed the different regulatory model and interaction between them, which proposes some valuable clues for the lncRNAs associated mechanisms in fiber development.
Similar content being viewed by others
Introduction
Cotton (Gossypium spp.) is one of the most important cash crops in the world because its main product fiber is the important natural source for the textile industry. In the four cultivars of Gossypium genus (G.hirsutum, G. barbadense, G. arboreum, and G.raimondii), G hirsutum (upland cotton) is the most widely planted due to its high yields and adaptability [42]. The period of cotton fiber development has been classified into four stages: initiation, elongation, secondary cell wall deposition, and maturity of fiber [12]. The first two stages could determine the number and length of fibers, further affecting fiber yields. Consequently, many studies have been documented to explore the underlying genetic mechanisms related to fiber initiation and elongation, contributing to cotton production improvement [16, 17, 19, 29, 65].
Cotton mutants with fibreless, fuzzless, and lintless are good materials for studying the mechanism of fiber initiation development. With the auxin and gibberellin (GA) application in two fibreless mutants of Asian cotton in vitro culture, it showed that fiber cells differentiated from ovule epidermis at a temperature lower than 30 degrees, but not above 32 degrees, which indicated the important roles of auxin and GA in fiber development promotion at some specific conditions [2]. SNPs comparison obtained by RNA-Seqs showed that glabrous mutant Xu142fl may be the progeny of G. barbadense. Based on the F2 and BC1 population between TM-1 and Xu142fl, the Li3 gene encoding an MYB-MIXTA-like transcription factor was mapped and adjacent to MYB25-like in the D12 chromosome [60]. The inheritance evaluation of fuzzless seed in segregation population suggested that the interaction of three loci (N1, n2 and n3) contributed to fuzzless seed [48], among which two loci, N1 and n2, located on a pair of homologous chromosomes A12/D12 [6]. The plants of N1N1 homozygous and N1n1 heterozygous produced fuzzless seeds [48]. The n3 locus that could produce the fibreless seed was identified by genetic analysis of cross progeny between N1N1 and n2n2 [48]. The fourth locus, named nt4\({n}_t^4\), was identified from ethyl methanesulfonate (EMS) induced mutation analysis, whose homozygous seed exhibited a partially naked phenotype [3]. All these fiber development defect mutants provide suitable materials for fiber development study.
With the advantage of Next Generation Sequencing (NGS), RNA-Seq as one of the NGS has been widely used to reveal expressions of genes and transcripts, among which some transcripts have been identified as non-coding RNA (ncRNA) because of their limitation of coding proteins. NcRNA includes microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and so on, which have emerged as key regulators of gene expression through their direct and indirect actions on chromatin [23,24,25]. In Oryza sativa, 1,254 differentially expressed lncRNAs (DELs) were identified from BIL progenies [26]. Another RNA-Seq showed that 328 of 444 DELs were associated with meiosis and the low fertility in autotetraploid rice [27]. The lncRNAs were also involved in abiotic stress such as drought and re-watering in Brassica napus [46], and osmotic and salt stress in Medicago truncatula [53]. The differences in genes expressions and regulations between fibreless mutants and wild-type have been investigated using omics methods [14, 28, 45, 51]. With fiberless mutant Xu142fl and its counterpart Xu142, a previous comparative small RNAome analysis uncovered a possible network of fiber initiation-related miRNAs in cotton ovules, which comprises seven miRNAs expressed in cotton ovules, and each of them bears functional specific targets [51]. Another work showed that 54 miRNAs are differentially expressed in fiber initiation between Xu142fl and its wild-type, which are potentially targeted to TFs such as MYB, auxin response factor, and Leucine repeat receptor [45]. Using multi-omics, the differentially expressed genes (1,953), proteins (187), and phosphoproteins (131) were identified by the comparison of Xu142 and Xu142fl [28]. Genetic markers including 302 SNPs for fiber development were also developed and validated based on a deep sequencing between Xu142 and Xu142fl [28]. In particular, a transcriptomic repertoire revealed that 645 and 651 lncRNAs were preferentially expressed in Xu142fl and Xu142, respectively. Further study showed that down-regulating two lncRNAs XLOC_545639 and XLOC_039050 in Xu142 fl increased the fiber initials on the ovules, while silencing XLOC_079089 in Xu142 shortened the fiber length [14], indicating the important and diverse roles of lncRNAs in fiber development.
LncRNA is a type of ncRNA with more than 200 nt in length and without protein-coding abilities [4, 51, 69]. Many studies on non-coding RNAs in cotton have been limited to small RNAs until now. For instance, a lot of miRNAs specifically expressed during anther development or callus were identified in male sterile cotton as well as cotton somatic embryogenesis [57, 64]. Gong et al. revealed the 33 conserved miRNAs families between the A and D genomes [9]. On the genomic level, the expression of 79 miRNAs families was studied and 257 novel miRNAs were identified related to cotton fiber elongation [63]. In addition, two key miR828 and miR858 were proved the roles in the regulation of homoeologous MYB2 (GhMYB2A and GhMYB2D) in G. hirsutum fiber development [11].
As a kind of long non-coding RNA, lncRNA provides more regulatory mechanisms for gene expression, protein synthesis, chromatin remodeling etc., while it is not clear about the detailed lncRNAs and the underlying mechanism in fiber development. A previous study identified 30,550 lincRNAs loci and 4,718 lncNATs loci, which are rich in repetitive sequences and preferentially expressed in a tissue-specific manner with weak evolutionary conservation. Further, lncRNAs showed overall higher methylation levels, and their expression was less affected by gene body methylation [52]. Using the epidermal cells from the ovules at 0 and 5 DPA from Xu142 and Xu142fl, 35,802 lncRNAs and 2,262 circular RNAs (circRNAs) were identified, of which 645 lncRNAs were preferentially expressed in the fibreless mutant Xu142fl and 651 lncRNAs were preferentially expressed in the fiber-attached lines; three lncRNAs XLOC_545639, XLOC_039050, and XLOC_079089 all showed the solid function in fiber development by VIGS assay [14]. Here, a novel glabrous mutant-ZM24fl, which showed excellent somatic embryogenesis induction was used to identify the key lncRNA involved in fiber initiation development [59].
Totally, 3,288 lncRNA transcripts were identified from the -2 DPA, 0 DPA and 5 DPA ovules of ZM24 and fl, which is significantly different from the number of identified lncRNA in Xu142fl [14] and G. barbadense L. cv 3-79 [52]. To identify the causal lncRNAs for fiber initiation, some comparisons were built to analyze the differentially expressed genes including lncRNAs and mRNAs during fiber initiation and earlier elongation. The identified DELs and DEGs in comparisons of 0 DPA vs -2 DPA and 5 DPA vs 0 DPA of ZM24 and fl indicated that many lncRNAs and coding genes are involved in the fiber initiation and primary development, while few lncRNAs and coding genes may involve the ovule development. The analysis of the DEGs further showed that fatty acid metabolism, very long strain fatty acid synthesis and sugar metabolism play important roles in the fiber initiation of ZM24, supporting the previous results [15, 39]. Moreover, some MYB family, bHLH type TFs encoding genes were also identified the important roles in fiber initiation, which is in agreement with the function of these TFs in previous research [10, 16, 29, 36, 40, 49, 50]. To uncover the upstream factors such as lncRNAs, we focused on the comparisons of ZM24_0 DPA vs fl_0 DPA and ZM24_0 DPA vs ZM24_-2 DPA to find the common lncRNAs which should be a key regulator for fiber initiation. Consequently, one lncRNA MSTRG 2723.1 was obtained, which locates on the A02G (84218766—84219942) encoding a lncNAT and covering the most coding region and partial 3’-terminal untranslated region of Ghicr24_A02G147600 (Figure S4). The co-expression analysis further identified its potent targets including 3-ketoacyl-CoA synthase, MYB family proteins, phosphatase 2C family proteins, pectin lysase, and some uncharacterized proteins, which may are involved in fiber initiation through fatty acid pathway, cell wall plasticity, MYB-mediated signaling etc. These results provide important clues for the upstream regulatory lncRNAs in fiber initiation and novel information associated with the fiber development regulation network. In addition, MSTRG 3390.1, MSTRG 48719.1, and MSTRG 31176.1 were also identified some positive correlation between fiber development and ovule development. The sequence analysis indicated that these lncRNAs are different from the previous lncRNAs XLOC_545639, XLOC_039050, and XLOC_079089 [14]. The target analysis also implied the possible interaction between different lncRNAs through mediating the common targets, which provide novel clues to explore the regulatory lncRNAs and underlying mechanisms in fiber development. Even with some achievement of lncRNAs, the understanding of the underlying mechanism of lncRNAs regulating targets or chromosome remodeling still needs more work to disclose.
Conclusion
Here, a novel glabrous cotton mutant ZM24fl was identified and applied to study the potential lncRNAs for fiber development with high-throughput sequencing. ZM24fl is derived from an elite cultivar of ZM24, which posses high callus induction and somatic embryogenesis ability, and is endowed with the valuable receptor for cotton genetic transformation [59]. Through the RNA-Seq and analysis in different ovules of ZM24 and fl, 3,288 lncRNAs were identified and some differentially expressed lncRNAs responsible for fiber (lint and fuzz) initiation and fiber earlier elongation were showed. Collectively, four lncRNAs MSTRG.2723.1, MSTRG.3390.1, MSTRG.48719.1 and MSTRG.31176.1 were showed potential important roles in fiber development, and the analysis of the target implied that MSTRG 2723.1 may function upstream of fatty acid metabolism, MBY25-mediating pathway, and pectin metabolism to regulate fiber initiation; the co-expression analysis between lncRNAs and targets further indicated the distinct models of different lncRNAs and interaction between lncRNAs, which provide precious information for illumination of the molecular mechanism of lncRNAs in fiber development of cotton.
Materials and methods
Plant Materials
Gossypium hirsutum L. acc. Zhongmiansuo24 (ZM24) and a natural fuzzless-lintless (fl) mutant from ZM24 were used and grown under standard field conditions in the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences (Zhengzhou research base, Henan). The ovule tissues were collected from cotton bolls on -2, 0, and 5 DPA using a sterile knife. All materials were frozen in liquid nitrogen immediately and stored at -80 °C for the following experiments.
Microscopic observation of fiber initiation on ovules epidermis
To study the fiber initiation phenotypes of ZM24 and fl, the cotton bolls of two lines on -2, 0, 1 and 2 DPA were collected. Then, the ovules were stripped from the bolls in the middle region. Immediate Scanning electron microscopy (Hitachi) was performed to observe the ovule epidermis as described previously [14].
Strand specific libraries construction and sequencing
Total RNAs of each ovule sample was extracted using the RNAprep Pure Plant Kit (Tiangen, Bei**g, China) following the manufacturer’s instruction. Total RNAs of each sample was quantified and qualified by Agilent 2100 Bio-analyzer (Agilent Technologies, Palo Alto, CA, USA), Nanodrop 2000 (Thermo Fisher Scientific Inc.), and 1% agarose gel. RNA with RIN value above 7 was used for following library construction. The rRNA was removed using the Ribo-Zero™ rRNA removal Kit. The ribosomal depleted RNA was then used for sequencing library preparation according to the manufacturer’s protocol (NEBNext® Ultra™ Directional RNA Library Prep Kit for Illumina®). The cDNA libraries with different indices were multiplexed and loaded on an Illumina Hiseq2500 with 150 base pair (bp) paired-end (PE150) raw reads according to the manufacturer’s instruction (Illumina, San Diego, CA, USA). RNA-Seq raw data with accession number SRP285346 was uploaded in the NCBI sequence read archive (http://www.ncbi.nlm.nih.gov/sra/) and the accession numbers of the twenty-four runs are SRR12710181-SRR12710192, and SRR12718970-SRR12718981.
Map** to the reference genome and LncRNAs identification
The raw data in fastq format were filtered with cutadapter (v1.9.1) software [30]. Clean data were obtained by removing reads that contained adapter, poly-N and base with Phred quality < 20 in 3’ or 5’ end, and the reads of length < 75 bp were removed after filtering. Finally, the GC percentage and Q30 of each sample were calculated using FastQC software (https://www.babraham.ac.uk/) and shown in Table S1. Clean data were mapped to the ZM24 genome (https://github.com/gitmalm/Genome-data-of-Gossypium-hirsutum/) [66] using HISAT(v2.1.0) [20, 21] software with the parameter “--rna-strandness RF”. Transcriptomes of each sample were assembled based on mapped reads and were merged by StringTie software (v2.0) [34, 35]. Transcripts annotation was performed using Cuffcompare [47]. Long non-coding RNA was identified as following steps: 1) transcripts with class codes of “i”, “u”, “x”, “j” representing the intronic transcripts, long intergenic noncoding RNAs (lincRNAs), long noncoding natural antisense transcripts (lncNAT), and the sense transcripts, respectively, were selected. 2) Transcripts with length > 200 bp, coverage > 1, FPKM > 0.5; 3) The CNCI [44], CPC [22] and PfamScan software were used to assessed protein-coding ability [7], with the parameter of (CPC score < 0, CNCI score < 0).
Differential expression analysis
The FPKM values and counts of genes and lncRNAs in each sample were calculated using StringTie and Ballgon [35]. Differential expression analyses were conducted by edgeR in R package [37, 38]. The DEGs and DELs were identified with an expression FPKM > 1.0, FDR (false discovery rate < 0.001), and |log2( fold change value)| ≥1 between each pairwise comparison.
Co-expression analysis between lncRNA and mRNA
To unveil the potential functions of DELs between the two genotypes, two interaction models of lncRNAs and protein-coding genes (lncRNAs/PC-genes) including cis- and trans-target were analyzed: 1) the Pearson correlation coefficient (PCC) between differentially expressed lncRNAs and mRNAs were calculated using the OmicShare tools (https://www.omicshare.com/) with the expression profiles (FPKM). The lncRNA-mRNA pairs with |PCC| > 0.95 and p-value < 0.01 were regarded as trans interaction between lncRNAs and mRNAs. 2) Protein-coding genes with a distance less than 20 kb from the upstream or downstream of lncRNAs were putative cis interaction. The co-expression networks were visualized by Cytoscape 3.6.1 [41].
GO and KEGG
To explore the functions of DEGs and lncRNAs between ZM24 and fl, the gene ontology (GO) enrichment was performed using the BLASTP program [1] and GO databases (http://archive.geneontology.org/latest-lite/) and (http://ftp.ncbi.nlm.nih.gov/gene/DATA/). Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed at KOBAS 3.0 website [58, 61] (http://kobas.cbi.pku.edu.cn/kobas3).
Q -PCR analysis
Ovules from bolls at -2, 0, 2, and 5 DPA were collected, and then total RNAs were extracted using the RNAprep Pure Plant Kit (Polysaccharides & Polyphenolics-rich, Tiangen, Bei**g, China) following the manufacturer’s instruction. Each reverse-transcribed reaction was performed with 1 μg RNA using a transScript® First-Strand cDNA Synthesis SuperMix (AT301-02, TransGen). The real-time PCR was performed on Roche 480 PCR system with a SYBR-Green Real-time PCR SuperMix (AQ101-01, TransGen). The 20 uL reaction volumes in each well contain 1 μL cDNA, 8.2 μL sterile water, 10 μL Mix, and 0.4 μL each of the forward and reverse primers. The Q-PCR procedures were as: pre-incubation of 30 s at 95 °C; followed by denaturation at 95 °C for 10 s, primer annealing at 55 °C for 10 s, and then extension at 72 °C for 30 s; finally, a melting curve at 95 °C for 30 s to check the primer specificity. The GhHistone3 (AF024716) gene was used as a reference gene. The 2-∆Ct method was used to calculate the relative expression of each gene, with three technical repetitions and three biological repetitions. Data were shown as mean ± SD. The student’s t-test was used for the significance statistic. The primer sequences used in the presented study are listed in Additional file 9.
Availability of data and materials
All the related data and files are presented including the sequences of the primers used in the Q-PCR. RNA-Seq raw data with accession number SRP285346 was uploaded in the NCBI sequence read archive (http://www.ncbi.nlm.nih.gov/sra/) and can be accessible with bioproject archive number PRJNA665585 (http://www.ncbi.nlm.nih.gov/bioproject/).
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Molecular biology. 1990;215(3):403–10. https://doi.org/10.1016/S0022-2836(05)80360-2.
Beasley CA. Temperature-dependent Response to Indoleacetic Acid Is Altered by NH (4) in Cultured Cotton Ovules. Plant physiol. 1977;59(2):203–6. https://doi.org/10.1104/pp.59.2.203.
Bechere E, Turley RB, Auld DL, Zeng LH. A New Fuzzless Seed Locus in an Upland Cotton (Gossypium hirsutum L.) Mutant. Am J Plant Sci. 2012;3(6):799–804. https://doi.org/10.4236/ajps.2012.36096.
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes & development. 2011;25(18):1915–27. https://doi.org/10.1101/gad.17446611.
Chekanova JA. Long non-coding RNAs and their functions in plants. Curr Opin Plant Biol. 2015;27:207–16. https://doi.org/10.1016/j.pbi.2015.08.003.
Endrizzi JE, Ramsay G. Identification of ten chromosome deficiencies of cotton. J Heredity. 1980;1:1. https://doi.org/10.1093/oxfordjournals.jhered.a109309.
Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85. https://doi.org/10.1093/nar/gkv1344.
Ge X, Zhang C, Wang Q, Yang Z, Wang Y, Zhang X, et al. iTRAQ protein profile differential analysis between somatic globular and cotyledonary embryos reveals stress, hormone, and respiration involved in increasing plantlet regeneration of Gossypium hirsutum L. J Proteome Res. 2015;14(1):268–78. https://doi.org/10.1021/pr500688g.
Gong L, Kakrana A, Arikit S, Meyers BC, Wendel JF. Composition and expression of conserved microRNA genes in diploid cotton (Gossypium) species. Genome Biol Evol. 2013;5(12):2449–59. https://doi.org/10.1093/gbe/evt196.
Guan XY, Li QJ, Shan CM, Wang S, Mao YB, Wang LJ, et al. The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiologia plantarum. 2008;134(1):174–82. https://doi.org/10.1111/j.1399-3054.2008.01115.x.
Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Comm. 2014;5:3050. https://doi.org/10.1038/ncomms4050.
Haigler CH, Betancur L, Stiff MR, Tuttle JR. Cotton fiber: a powerful single-cell model for cell wall and cellulose research. Front Plant Sci. 2012;3:104. https://doi.org/10.3389/fpls.2012.00104.
Hoffmann L, Besseau S, Geoffroy P, Ritzenthaler C, Meyer D, Lapierre C, et al. Silencing of hydroxycinnamoyl-coenzyme A shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell. 2004;16(6):1446–65. https://doi.org/10.1105/tpc.020297.
Hu H, Wang M, Ding Y, Zhu S, Zhao G, Tu L, et al. Transcriptomic repertoires depict the initiation of lint and fuzz fibres in cotton (Gossypium hirsutum L.). Plant Biotechnol J. 2018;16(5):1002–12. https://doi.org/10.1111/pbi.12844.
Hu W, Chen L, Qiu X, Wei J, Shen G. AKR2A participates in the regulation of cotton fiber development by modulating biosynthesis of very ong hain fatty acids. Plant Biotechnol J. 2019;18(2). https://doi.org/10.1111/pbi.13221.
Huang Y, Liu X, Tang K, Zuo K. Functional analysis of the seed coat-specific gene GbMYB2 from cotton. Plant Physiol Biochem. 2013;73:16–22. https://doi.org/10.1016/j.plaphy.2013.08.004.
Huang G, Huang JQ, Chen XY, Zhu YX. Recent Advances and Future Perspectives in Cotton Research. Annu Rev Plant Biol. 2021. https://doi.org/10.1146/annurev-arplant-080720-113241.
Humphries JA, Walker AR, Timmis JN, Orford SJ. Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant molecular biology. 2005;57(1):67–81. https://doi.org/10.1007/s11103-004-6768-1.
Suo J, Liang X, Pu L, Zhang Y, Xue Y. Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochimica et Biophysica Acta (BBA). Gene Structure and Expression. 2003;1630(1):25–34. ISSN 0167-4781. https://doi.org/10.1016/j.bbaexp.2003.08.009.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–60. https://doi.org/10.1038/nmeth.3317.
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genoty** with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37(8):907–15. https://doi.org/10.1038/s41587-019-0201-4.
Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, et al. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic acids research. 2007;35(Web Server issue):W345–9. https://doi.org/10.1093/nar/gkm391.
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC biology. 2013;11:59. https://doi.org/10.1186/1741-7007-11-59.
Kowalczyk MS, Higgs DR, Gingeras TR. RNA discrimination. Nature. 2012;482(7385):310–1. https://doi.org/10.1038/482310a.
Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–69. https://doi.org/10.1534/genetics.112.146704.
Li M, Cao A, Wang R, Li Z, Li S, Wang J. Genome-wide identification and integrated analysis of lncRNAs in rice backcross introgression lines (BC (2) F (12)). BMC plant biology. 2020a;20(1):300. https://doi.org/10.1186/s12870-020-02508-y.
Li X, Shahid MQ, Wen M, Chen S, Yu H, Jiao Y, et al. Global identification and analysis revealed differentially expressed lncRNAs associated with meiosis and low fertility in autotetraploid rice. BMC plant biology. 2020b;20(1):82. https://doi.org/10.1186/s12870-020-2290-0.
Ma Q, Wu M, Pei W, Wang X, Zhai H, Wang W, et al. RNA-Seq-Mediated Transcriptome Analysis of a Fiberless Mutant Cotton and Its Possible Origin Based on SNP Markers. PloS one. 2016;11(3):e0151994. https://doi.org/10.1371/journal.pone.0151994.
Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J. 2009;59(1):52–62. https://doi.org/10.1111/j.1365-313X.2009.03847.x.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–2. https://doi.org/10.14806/ej.17.1.200.
Mattick JS, Rinn JL. Discovery and annotation of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):5–7. https://doi.org/10.1038/nsmb.2942.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Gen. 2009;10(3):155–9. https://doi.org/10.1038/nrg2521.
Millar AA, Clemens S, Zachgo S, Giblin EM, Taylor DC, Kunst L. CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell. 1999;11(5):825–38. https://doi.org/10.1105/tpc.11.5.825.
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–5. https://doi.org/10.1038/nbt.3122.
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–67. https://doi.org/10.1038/nprot.2016.095.
Pu L, Li Q, Fan X, Yang W, Xue Y. The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics. 2008;180(2):811–20. https://doi.org/10.1534/genetics.108.093070.
Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–7. https://doi.org/10.1093/bioinformatics/btm453.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. https://doi.org/10.1093/bioinformatics/btp616.
Ruan YL, Llewellyn DJ, Furbank RT, Chourey PS. The delayed initiation and slow elongation of fuzz-like short fibre cells in relation to altered patterns of sucrose synthase expression and plasmodesmata gating in a lintless mutant of cotton. J Exp Bot. 2005;56(413):977–84. https://doi.org/10.1093/jxb/eri091.
Shan C-M, Shangguan X-X, Zhao B, Zhang X-F, Chao L-M, Yang C-Q, et al. Control of cotton fibre elongation by a homeodomain transcription factor GhHOX3. Nat Comm. 2014;5:5519. https://doi.org/10.1038/ncomms6519.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003;13(11):2498–504. https://doi.org/10.1101/gr.1239303.
Shao Q, Zhang F, Tang S, Liu Y, Fang X, Liu D, et al. Identifying QTL for fiber quality traits with three upland cotton (Gossypium hirsutum L.) populations. Euphytica. 2014;198(1):43–58. https://doi.org/10.1007/s10681-014-1082-8.
Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, et al. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J Ex Bot. 2014;65(17):4975–83. https://doi.org/10.1093/jxb/eru256.
Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, et al. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic acids research. 2013;41(17):e166. https://doi.org/10.1093/nar/gkt646.
Sun R, Li C, Zhang J, Li F, Ma L, Tan Y, et al. Differential expression of microRNAs during fiber development between fuzzless-lintless mutant and its wild-type allotetraploid cotton. Scientific reports. 2017;7(1):3. https://doi.org/10.1038/s41598-017-00038-6.
Tan X, Li S, Hu L, Zhang C. Genome-wide analysis of long non-coding RNAs (lncRNAs) in two contrasting rapeseed (Brassica napus L.) genotypes subjected to drought stress and re-watering. BMC plant biology. 2020;20(1):81. https://doi.org/10.1186/s12870-020-2286-9.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. https://doi.org/10.1038/nbt.1621.
Turley RB, Kloth RH. Identification of a third fuzzless seed locus in upland cotton (Gossypium hirsutum L.). J Heredity. 2002;93(5):359–64. https://doi.org/10.1093/jhered/93.5.359.
Walford SA, Wu Y, Llewellyn DJ, Dennis ES. GhMYB25-like: a key factor in early cotton fibre development. Plant J. 2011;65(5):785–97. https://doi.org/10.1111/j.1365-313X.2010.04464.x.
Walford SA, Wu Y, Llewellyn DJ, Dennis ES. Epidermal cell differentiation in cotton mediated by the homeodomain leucine zipper gene, GhHD-1. Plant J. 2012;71(3):464–78. https://doi.org/10.1111/j.1365-313X.2012.05003.x.
Wang ZM, Xue W, Dong CJ, ** LG, Bian SM, Wang C, et al. A comparative miRNAome analysis reveals seven fiber initiation-related and 36 novel miRNAs in develo** cotton ovules. Molecular plant. 2012;5(4):889–900. https://doi.org/10.1093/mp/ssr094.
Wang M, Yuan D, Tu L, Gao W, He Y, Hu H, et al. Long noncoding RNAs and their proposed functions in fibre development of cotton (Gossypium spp.). New phytologist. 2015a;207(4):1181–97. https://doi.org/10.1111/nph.13429.
Wang TZ, Liu M, Zhao MG, Chen R, Zhang WH. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC plant biology. 2015b;15:131. https://doi.org/10.1186/s12870-015-0530-5.
Wang XC, Li Q, ** X, **ao GH, Liu GJ, Liu NJ, et al. Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. J Proteomics. 2015c;114:16–27. https://doi.org/10.1016/j.jprot.2014.10.022.
Wang M, Zhao W, Gao L, Zhao L. Genome-wide profiling of long non-coding RNAs from tomato and a comparison with mRNAs associated with the regulation of fruit ripening. BMC plant biology. 2018;18(1):75. https://doi.org/10.1186/s12870-018-1300-y.
Wang Z, Yang Z, Li F. Updates on molecular mechanisms in the development of branched trichome in Arabidopsis and nonbranched in cotton. Plant Biotechnol J. 2019;17(9). https://doi.org/10.1111/pbi.13167.
Wei M, Wei H, Wu M, Song M, Zhang J, Yu J, et al. Comparative expression profiling of miRNA during anther development in genetic male sterile and wild type cotton. BMC plant biology. 2013;13:66. https://doi.org/10.1186/1471-2229-13-66.
Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic acids research. 2006;34(Web Server issue):W720–4. https://doi.org/10.1093/nar/gkl167.
Wu X, Li F, Zhang C, Liu C, Zhang X. Differential gene expression of cotton cultivar CCRI24 during somatic embryogenesis. J Plant Physiol. 2009;166(12):1275–83. https://doi.org/10.1016/j.jplph.2009.01.012.
Wu H, Tian Y, Wan Q, Fang L, Guan X, Chen J, et al. Genetics and evolution of MIXTA genes regulating cotton lint fiber development. New Phytologist. 2018;217(2):883–95. https://doi.org/10.1111/nph.14844.
**e C, Mao X, Huang J, Ding Y, Wu J, Dong S, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39(Web Server issue):W316–22. https://doi.org/10.1093/nar/gkr483.
**n M, Wang Y, Yao Y, Song N, Hu Z, Qin D, et al. Identification and characterization of wheat long non-protein coding RNAs responsive to powdery mildew infection and heat stress by using microarray analysis and SBS sequencing. BMC plant biol. 2011;11:61. https://doi.org/10.1186/1471-2229-11-61.
Xue W, Wang Z, Du M, Liu Y, Liu JY. Genome-wide analysis of small RNAs reveals eight fiber elongation-related and 257 novel microRNAs in elongating cotton fiber cells. BMC genomics. 2013;14:629. https://doi.org/10.1186/1471-2164-14-629.
Yang J, Liu X, Xu B, Zhao N, Yang X, Zhang M. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC genomics. 2013;14:9. https://doi.org/10.1186/1471-2164-14-9.
Yang Z, Zhang C, Yang X, Liu K, Wu Z, Zhang X, et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytologist. 2014;203(2):437–48. https://doi.org/10.1111/nph.12824.
Yang Z, Ge X, Yang Z, Qin W, Sun G, Wang Z, et al. Extensive intraspecific gene order and gene structural variations in upland cotton cultivars. Nat Comm. 2019;10(1):2989. https://doi.org/10.1038/s41467-019-10820-x.
Zhang YC, Chen YQ. Long noncoding RNAs: new regulators in plant development. Biochem Biophys Res Commun. 2013;436(2):111–4. https://doi.org/10.1016/j.bbrc.2013.05.086.
Zhang L, Wang M, Li N, Wang H, Qiu P, Pei L, et al. Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol J. 2018;16(6):1172–85. https://doi.org/10.1111/pbi.12861.
Zhao T, Xu X, Wang M, Li C, Li C, Zhao R, et al. Identification and profiling of upland cotton microRNAs at fiber initiation stage under exogenous IAA application. BMC genomics. 2019;20(1):421. https://doi.org/10.1186/s12864-019-5760-8.
Zhou ZY, Li AM, Adeola AC, Liu YH, Irwin DM, **e HB, et al. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biol Evol. 2014;6(6):1387–92. https://doi.org/10.1093/gbe/evu113.
Acknowledgments
We thank Yong Cheng and **n Li (Zhengzhou Research Base, Institute of Cotton Research of CAAS) for technical assistance.
Funding
This study was supported financially by the National Natural Science Foundation of China (32072022 and 31690093), the Creative Research Groups of China (31621005), and Central Public-interest Scientific Institution Basal Research Fund (1610162020010202) for scientific research into non-profit industries.
Author information
Authors and Affiliations
Contributions
Zhi Wang designed the study, **anyan Zou and Faiza Ali wrote the main manuscript text and prepared all figures. Shuangxia ** and Fuguang Li edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
RNA-Seq data for 12 samples
Additional file 2.
List of lncRNAs identified from ovules of two lines during fiber initiation stage.
Additional file 3.
Differentially expressed lncRNAs in different comparisons.
Additional file 4.
Differentially expressed genes in different comparisons.
Additional file 5.
The targeted genes by lncRNA MSTRG.2723.1.
Additional file 6.
Transcription factors identification in the 12,971 DEGs.
Additional file 7.
Differentially expressed lncRNA and their targets in all comparisons.
Additional file 8.
The KEGG pathways of differentially expressed targeted genes of DELs.
Additional file 9.
List of primers used in this research.
Additional file 10: Figure S1.
Observation and comparison of fl and ZM24 in the different developmental stages and tissues. a and f The plants architecture of fl and ZM24; b and c The magnifications of white rectangular dotted bolls at 20 DPA from a and f, respectively. d The size comparison of bolls at 15 DPA from fl (left) and ZM24 (right). e The size and shape of leaves from two lines fl (left) and ZM24 (right). g and iThe epidermal hair on the abaxial leaf surface of fl and ZM24; (h) The stem epidermal hair of fl (up) and ZM24 (down); bars in a-f: 2.0 cm; bars in g-i: 1000 μm.
Additional file 11: Figure S2.
Heatmap shows the Pearson correlation coefficients among the 12 samples.
Additional file 12: Figure S3.
Gene ontology classifications of DEGs in ovules of ZM24 vs fl at 5 PDA. The most highly enriched GO terms showed the 1,378 down- and 2,608 up-regulated genes in ovules of ZM24 vs fl at 5 DPA.
Additional file 13: Figure S4.
The physical location of lncRNA MSTRG.2723.1 on the ZM24 genome. The MSTRG.2723.1 is a natural antisense transcript and overlaps with the gene of Ghicr24_A02G147600. Blue and orange rectangles represent exons and introns, respectively. Arrows indicate the direction of transcription.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zou, X., Ali, F., **, S. et al. RNA-Seq with a novel glabrous-ZM24fl reveals some key lncRNAs and the associated targets in fiber initiation of cotton. BMC Plant Biol 22, 61 (2022). https://doi.org/10.1186/s12870-022-03444-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-022-03444-9