Abstract
Background
Flesh is prone to accumulate more anthocyanin in postharvest ‘Friar’ plum (Prunus salicina Lindl.) fruit stored at an intermediate temperature. However, little is known about the molecular mechanism of anthocyanin accumulation regulated by storage temperature in postharvest plum fruit.
Results
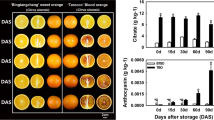
To reveal the potential molecular regulation mechanism of anthocyanin accumulation in postharvest ‘Friar’ plum fruit stored at different temperatures (0 °C, 10 °C and 25 °C), the fruit quality, metabolite profile and transcriptome of its flesh were investigated. Compared to the plum fruit stored at 0 °C and 25 °C, the fruit stored at 10 °C showed lower fruit firmness after 14 days and reduced the soluble solids content after 21 days of storage. The metabolite analysis indicated that the fruit stored at 10 °C had higher contents of anthocyanins (pelargonidin-3-O-glucoside, cyanidin-3-O-glucoside, cyanidin-3-O-rutinoside and quercetin-3-O-rutinose), quercetin and sucrose in the flesh. According to the results of weighted gene coexpression correlation network analysis (WGCNA), the turquoise module was positively correlated with the content of anthocyanin components, and flavanone 3-hydroxylase (F3H) and chalcone synthase (CHS) were considered hub genes. Moreover, MYB family transcription factor APL (APL), MYB10 transcription factor (MYB10), ethylene-responsive transcription factor WIN1 (WIN1), basic leucine zipper 43-like (bZIP43) and transcription factor bHLH111-like isoform X2 (bHLH111) were closely related to these hub genes. Further qRT–PCR analysis verified that these transcription factors were specifically more highly expressed in plum flesh stored at 10 °C, and their expression profiles were significantly positively correlated with the structural genes of anthocyanin synthesis as well as the content of anthocyanin components. In addition, the sucrose biosynthesis-associated gene sucrose synthase (SS) was upregulated at 10 °C, which was also closely related to the anthocyanin content of plum fruit stored at 10 °C.
Conclusions
The present results suggest that the transcription factors APL, MYB10, WIN1, bZIP43 and bHLH111 may participate in the accumulation of anthocyanin in ‘Friar’ plum flesh during intermediate storage temperatures by regulating the expression of anthocyanin biosynthetic structural genes. In addition, the SS gene may play a role in anthocyanin accumulation in plum flesh by regulating sucrose biosynthesis.
Similar content being viewed by others
Background
Anthocyanin is a kind of water-soluble flavonoid that is derived from the branch of flavonoids, and it gives flowers and fruits various and graceful colours [1, 2]. As an antioxidant, anthocyanin can effectively remove free radicals such as reactive oxygen species (ROS) when plants suffer environmental stress, protecting plants from damage [3]. In addition, it has been shown that anthocyanin intake is beneficial to human prevention of cardiovascular diseases and cancer [4]; thus, anthocyanin has been widely studied recently.
The biosynthetic pathway of anthocyanins in higher plants is conserved, and anthocyanins are synthesized from phenylalanine catalysed by a series of enzymes. The enzyme-associated genes involved in anthocyanin synthesis are divided into early biosynthesis genes (EBGs) and late biosynthesis genes (LBGs) [5]. EBGs include chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H) and flavonoid-3′-hydroxylase (F3’H), and they are common to different flavonoid synthesis branches [6, 7]. LBGs mainly include dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase/anthocyanin synthetase (LDOX/ANS) and UDP-glucose: flavonoid 3-O-glucosyltransferase (UFGT), and they contribute to the production of various anthocyanin components by catalysing flavanonol and its subsequent derivatives [8]. Anthocyanin biosynthesis-related genes are regulated by many transcription factors, among which MYB-bHLH-WD40 (MBW) has been widely studied. The MBW complex positively regulates the expression of structural genes by binding to cis-acting elements on the promoter regions of genes (such as DFR, LDOX/ANS, UFGT, etc.) and then facilitates the accumulation of anthocyanin in plants [7, 9,10,11,12]. In addition, transcription factors such as COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1), JAZ (JASMONATE ZIM-DOMAIN), NAC (NAM, ATAF1/2, CUC2), SPL (SQUAMOSA promoter-binding protein-like) and WRKY have been considered to regulate anthocyanin biosynthesis by interacting with the MBW complex [13,14,15,16,17,18].
Anthocyanin accumulation can be affected by light, temperature, hormones and mineral nutrition, and favourable low-temperature conditions are one of the important factors that induce the biosynthesis of anthocyanins [16, 19,20,21,22,3a). To screen the candidate genes related to anthocyanin biosynthesis, our study mainly focused on the DEGs at 10 °C vs. 0 °C and 10 °C vs. 25 °C. There were 2807, 4008, 4450, and 4234 genes that were differentially expressed on Days 7, 14, 21, and 28, respectively, at 10 °C vs. 0 °C and 10 °C vs. 25 °C (Fig. 3b). Among them, 467 genes were consistently differentially expressed (Fig. 3b).
Summary of differentially expressed genes under different storage temperatures in ‘Friar’ plum. a Number of DEGs in different DEG sets. T0–7 vs. T10–7 represents the DEG set in which samples stored at 0 °C for 7 days versus samples stored at 10 °C for 7 days. b Venn diagram shows DEGs in both 10 °C vs. 0 °C and 10 °C vs. 25 °C at different storage time points. DAT represents days after treatment
Construction of a WGCNA and coexpression network
To obtain hub genes related to anthocyanin accumulation, the relationships of DEGs, anthocyanin components and storage temperature for each sample were analysed by constructing a WGCNA (Fig. 4). Sample clustering showed that the three biological replicates of each treatment were very good (Fig. 4a). Ten coexpression modules were identified by WGCNA (Fig. 4b), among which the turquoise module was positively correlated with the contents of pelargonidin-3-O-glucoside (r = 0.67, p value = 3e-06), cyanidin-3-O-glucoside (r = 0.80, p value = 8e-11), cyanidin-3-O-rutinoside (r = 0.75, p value = 4e-08), and quercetin-3-O-rutinose (r = 0.83, p value = 6e-11). In addition, the turquoise module was positively correlated with the storage temperature of 10 °C, and the correlation coefficient was 0.90 (p value = 6e-15) (Fig. 4c). According to GO and KEGG enrichment of the candidate genes in the turquoise module (1416 genes in total), 33 genes were mapped to the flavonoid metabolism pathway, and 31 genes were mapped to the starch and sugar metabolism pathway (see Additional file 1: Figs. S1 and S2).
Weighted gene coexpression network analysis of ‘Friar’ plum fruit under different storage temperatures. a Sample clustering. A01–03 (initial sample); B01–03, C01–03, D01–03 (sample stored at 0 °C, 10 °C, and 25 °C for 7 days, respectively); B04–06, C04–06, D04–06 (sample stored at 0 °C, 10 °C, and 25 °C for 14 days), B07–09, C07–09, D07–09 (sample stored at 0 °C, 10 °C, and 25 °C for 21 days), B10–12, C10–12, D10–12 (sample stored at 0 °C, 10 °C, and 25 °C for 28 days). b Hierarchical clustering showing modules of coexpressed genes. c Module/trait correlations and corresponding p values. The right panel shows a colour scale for module/trait correlations from −1 to 1
Identification of candidate genes involved in anthocyanin biosynthesis
A total of 43 structural genes involved in anthocyanin biosynthesis were obtained in the turquoise module, and a heatmap of their expression profiles in the flesh of ‘Friar’ plum fruit was drawn based on their FPKM value (log10(FPKM+1)) (Fig. 5). The 43 structural genes from all major steps of the anthocyanin biosynthesis pathway were distributed as follows: four phenylalanine ammonia-lyase genes (PAL), one 4-coumarate: coenzyme A ligase (4CL), fourteen chalcone synthase genes (CHS), four chalcone isomerase genes (CHI), one flavonoid-3′-hydroxylase gene (F3’H), four flavanone 3-hydroxylase genes (F3H), three dihydroflavonol 4-reductase genes (DFR), five leucoanthocyanidin dioxygenase/anthocyanin synthase genes (LDOX/ANS) and seven UDP-glucose: flavonoid 3-O-glucosyltransferase genes (UFGT).
Analysis of genes related to anthocyanin biosynthesis in the turquoise module. a Anthocyanin biosynthetic pathway. The bold font indicates the genes obtained in the turquoise module and the anthocyanin components detected in the flesh of ‘Friar’ plum fruit. b Heatmap of the expression levels of differentially expressed genes (DEGs) involved in anthocyanin biosynthesis. c qRT–PCR detection of anthocyanin synthesis-related structural genes, PsPAL (Pd.00 g835470), PsCHS (Pd.00 g300780), PsCHI (Pd.00 g402750), PsF3H (Pd.00 g891590), PsF3’H (Pd.00 g637710), PsDFR (Pd.00 g1089860), PsLDOX (Pd.00 g746630), and PsUFGT (Pd.00 g247850). d Correlation analysis of the expression profiles in qRT–PCR (qPCR) and transcriptome data (RNA-seq) (N = 13, |r| > 0.55 represents a significant correlation between the two sets of data)
Furthermore, the expression patterns of eight representative structural genes involved in anthocyanin biosynthesis, PsPAL (Pd.00 g835470), PsCHS (Pd.00 g300780), PsCHI (Pd.00 g402750), PsF3H (Pd.00 g891590), PsF3’H (Pd.00 g637710), PsDFR (Pd.00 g1089860), PsLDOX (Pd.00 g746630) and PsUFGT (Pd.00 g247850), were studied via qRT–PCR, and the transcripts of these genes were significantly higher in the flesh of plum fruit stored at 10 °C than in that stored at 0 °C and 25 °C (Fig. 5c), which was consistent with the results of transcriptome analysis based on the correlation analysis (Fig. 5d).
Identification of genes involved in carbohydrate metabolism
Carbohydrates are considered substrates for anthocyanin synthesis, and the change in soluble sugar content was detected by HPLC. The contents of glucose, fructose and sorbitol showed downward trends at three different storage temperatures. The sucrose content decreased in the flesh of plum fruit during 0 °C storage but increased during 10 °C and 25 °C storage; moreover, it was higher at 0 °C than at 25 °C (Fig. 6a). In the turquoise module, which was related to anthocyanin synthesis, five genes were involved in starch and sugar metabolism, including two hexokinases (HXKs) and three sucrose synthases (SSs) (Fig. 6b). Correlation analysis showed that the expression patterns of the SS genes were positively correlated with sucrose content and anthocyanin content (Fig. 6c), suggesting that higher expression levels of these genes were beneficial to carbohydrate metabolism, which contributed to anthocyanin accumulation in the flesh under storage at 10 °C.
Soluble sugar content and expression pattern of carbohydrate metabolism-related genes under different storage temperatures. a Glucose, fructose, sorbitol and sucrose contents in ‘Friar’ plum fruit under different storage temperatures. b Heatmap of the expression levels of carbohydrate metabolism-related genes in the turquoise module. c Correlation analysis of anthocyanin content, soluble sugar content and expression profiles of carbohydrate metabolism-related genes (N = 13, |r| > 0.55 represents a significant correlation between the two sets of data). Different lowercase letters above the bars indicate significant differences (P < 0.05), which were obtained based on one-way ANOVA by LSD and DUNCAN tests
Screening potential transcription factors that regulate anthocyanin synthesis
To further explore the molecular regulatory mechanism of anthocyanin biosynthesis in the flesh of ‘Friar’ plum fruit, a coexpression network was constructed based on the genes present in the turquoise module. In the network, three F3H genes (Pd.00 g799840, Pd.00 g891590 and Pd.00 g617550) and two CHS genes (Pd.00 g276460 and Pd.00 g113960) were identified as hub genes (Fig. 7a), and five transcription factor genes, MYB10, APL, WIN1, bHLH111 and bZIP43, were found to be coexpressed with anthocyanin biosynthesis-related genes. Among the five transcription factor genes, MYB10 and WIN1 were closely related to CHS, F3H, DFR and LDOX, APL was closely related to CHS, F3H and DFR, bHLH111 was only closely related to F3H, and bZIP43 was closely related to CHS and F3H. In addition, there was a positive correlation between APL and WIN1 (Fig. 7b).
Coexpression network analysis of the potential key genes in turquoise modules based on WGCNA. a Coexpression network of the genes (weight > 0.5). b Coexpression network of transcription factors and anthocyanin biosynthesis-related structural genes. c Heatmap of the expression levels of the candidate transcription factors. d qRT–PCR detection of candidate transcription factor genes PsAPL (Pd.00 g301130), PsWIN1 (Pd.00 g307540), PsMYB10 (Pd.00 g623010), PsbHLH111 (Pd.00 g1033540), and PsbZIP43 (Pd.00 g297380). e Correlation analysis of the expression profiles in qRT–PCR (qPCR) and transcriptome data (RNA-seq) (N = 13, |r| > 0.55 represents a significant correlation between the two sets of data)
The heatmap based on the transcriptome data showed that the expression levels of APL, WIN1, MYB10, bHLH111 and bZIP43 were obviously upregulated in the fruit stored at 10 °C, while there was little change in the fruit stored at 0 °C and 25 °C (Fig. 7c). Meanwhile, the qRT–PCR results indicated that the transcripts of these five detected genes in the fruit stored at 10 °C were markedly higher than those in the fruit stored at 0 °C and 25 °C (Fig. 7d), which was consistent with the transcriptome analysis results based on the correlation analysis (Fig. 7e). Furthermore, the expression patterns of the five transcription factor genes were positively correlated with the changes in the expression levels of the structural genes and the anthocyanin content (see Additional file 1: Fig. S3). It was proposed that APL, WIN1, MYB10, bHLH111 and bZIP43 might be involved in anthocyanin accumulation by regulating the expression of structural genes associated with anthocyanin biosynthesis.
Discussion
Intermediate temperature is beneficial to promote anthocyanin accumulation in plum fruit
Temperature is an important environmental factor that affects anthocyanin accumulation. Low temperature (4 °C) can significantly induce anthocyanin accumulation in Arabidopsis seedlings in the presence of light [24]. Intermediate temperature (16 °C) effectively leads to reddening in the leaves of apple and begonia [29, 30], and 15 °C treatment promotes anthocyanin accumulation in grape peel [31]. For postharvest fruits, intermediate temperature also promotes the process of anthocyanin accumulation in peach, kiwifruit, sweet orange and plum fruits [26,27,28, 32,66] to obtain mapped data, and evaluated for library quality, such as insert length tests and randomness tests. Then, structural-level analysis, such as alternative splicing analysis, new gene discovery and gene structure optimization, was carried out. The expression levels of differentially expressed genes, functional annotation and functional enrichment of differentially expressed genes were analysed. Finally, the transcriptome data and phenotypic data were analysed by WGCNA using R language.
RNA extraction and qPCR analysis
Total RNA from flesh was extracted by the CTAB method [67]. RNA (0.8 μg, OD260:OD280 between 1.80 and 2.0, OD260:OD230 > 1.5, no obvious degradation by electrophoresis) was used for reverse transcription by PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Biomedicals, Dalian, China). The products were diluted 15 times with nuclease-free water and then subjected to real-time fluorescence quantitative PCR (qPCR) with a TB Green® Premix Ex Taq™ II (Tli RNaseH Plus) kit (TaKaRa Biomedicals).
Quantitative real-time (qRT)-PCR assays were conducted using an Applied Biosystems 7500 Fast Real-Time PCR System. The reaction system was 20 μL, including 10 μL of 2X Green Premix Ex Taq II (Tli RNaseH Plus), 0.8 μL each of gene specific upstream primer and downstream primer, 0.4 μL of ROX Reference Dye II (50X), 2 μL of diluted cDNA, and 6 μL of nuclease-free water. The running program was set as follows: 30 s at 95 °C for one cycle, 5 s at 95 °C and 34 s at 60 °C for 40 cycles. PsACTIN7 was used as the internal reference. The primers used in this paper are listed in Additional file 1: Table S1. The relative expression levels of genes were calculated according to the 2-△△Ct method.
Statistical analysis
Each experiment was performed in three replicates. Experimental results were analysed using GraphPad Prism 8, Origin 2021, IBM SPSS Statistics 23, RStudio, and Cytoscape 3.7.1 software. Error bars denote standard deviations. Different lowercase letters above the bars indicate significant differences (P < 0.05), which were obtained based on one-way ANOVA by LSD and DUNCAN tests using IBM SPSS Statistics 23 software.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its additional file(s).
Abbreviations
- WGCNA:
-
Weighted gene co-expression correlation network analysis
- ROS:
-
Reactive oxygen species
- EBGs:
-
Early biosynthesis genes
- LBGs:
-
Late biosynthesis genes
- PAL:
-
Phenylalnine ammonialyase
- CHS:
-
Chalcone synthase
- CHI:
-
Chalcone isomerase
- F3H:
-
Flavanone 3-hydroxylase
- F3’H:
-
Flavonoid-3′-hydroxylase
- DFR:
-
Dihydroflavonol 4-reductase
- LDOX/ANS:
-
Leucoanthocyanidin dioxygenase/Anthocyanin synthetase
- UFGT:
-
UDP-glucose: flavonoid 3-O-glucosyltransferase
- MBW:
-
MYB-bHLH-WD40
- COP1:
-
CONSTITUTIVE PHOTOMORPHOGENIC 1
- JAZ:
-
JASMONATE ZIM-DOMAIN
- NAC:
-
NAM, ATAF1/2, CUC2
- SPL:
-
SQUAMOSA promoter-binding protein-like
- HXK:
-
Hexokinase
- SS:
-
sucrose synthases
- APL:
-
MYB family transcription factor APL
- MYB10:
-
MYB10 transcription factor
- WIN1:
-
Ethylene-responsive transcription factor WIN1
- bZIP43:
-
Basic leucine zipper 43-like
- bHLH111:
-
Transcription factor bHLH111-like isoform X2
- LTR:
-
Low temperature response
- MdEIL1:
-
EIN3-LIKE1
- MdERF1b:
-
ETHYLENE RESPONSE FACTOR1b
- DAT:
-
Days after treatment
- SSC:
-
Soluble solids content
- FPKM:
-
Fragments per kilobase of exon model per million mapped fragments
- DEGs:
-
Differentially expressed genes
- UPLC–MS/MS:
-
ultra-performance liquid chromatography–tandem mass spectrometry
- HPLC:
-
High performance liquid chromatography
References
Bai S, Tao R, Yin L, Ni J, Yang Q, Yan X, et al. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia ELONGATED HYPOCOTYL 5 in the peel of pear fruit. Plant J. 2019;100:1208–23.
Davies KM, Albert NW, Schwinn KE. From landing lights to mimicry: the molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct Plant Biol. 2012;39:619–38.
Lev-Yadun S, Gould KS. Role of anthocyanins in plant defence. In: Winefield C, Davies K, Gould K, editors. Anthocyanins. New York: Springer; 2008. p. 22–8.
Tsuda T. Dietary anthocyanin-rich plants: biochemical basis and recent progress in health benefits studies. Mol Nutr Food Res. 2012;56:159–70.
Pelletier MK, Murrell JR, Shirley BW. Characterization of flavonol synthase and leucoanthocyanidin dioxygenase genes in Arabidopsis. Further evidence for differential regulation of "early" and "late" genes. Plant Physiol. 1997;113:1437–45.
Jeong SW, Das PK, Jeoung SC, Song JY, Lee HK, Kim YK, et al. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol. 2010;154:1514–31.
Ni J, Zhao Y, Tao R, Yin L, Gao L, Strid A, et al. Ethylene mediates the branching of the jasmonate-induced flavonoid biosynthesis pathway by suppressing anthocyanin biosynthesis in red Chinese pear fruits. Plant Biotechnol J. 2020;18:1223–40.
Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, et al. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol Biochem. 2013;72:21–34.
Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27.
Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–85.
Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34.
Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–42.
Gonzalez A, Brown M, Hatlestad G, Akhavan N, Smith T, Hembd A, et al. TTG2 controls the developmental regulation of seed coat tannins in Arabidopsis by regulating vacuolar transport steps in the proanthocyanidin pathway. Dev Biol. 2016;419:54–63.
Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22.
Maier A, Schrader A, Kokkelink L, Falke C, Welter B, Iniesto E, et al. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013;74:638–51.
Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, et al. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–814.
Verweij W, Spelt CE, Bliek M, de Vries M, Wit N, Faraco M, et al. Functionally similar WRKY proteins regulate vacuolar acidification in petunia and hair development in Arabidopsis. Plant Cell. 2016;28:786–803.
Zhou H, Lin-Wang K, Wang H, Gu C, Dare AP, Espley RV, et al. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015;82:105–21.
Bai S, Saito T, Moriguchi T, Honda C, Hatsuyama Y, Ito A. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta. 2014;240:1051–62.
Crifo T, Puglisi I, Petrone G, Recupero GR, Lo Piero AR. Expression analysis in response to low temperature stress in blood oranges: implication of the flavonoid biosynthetic pathway. Gene. 2011;476:1–9.
Das PK, Shin DH, Choi SB, Park YI. Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cell. 2012;34:501–7.
Feng S, Wang Y, Yang S, Xu Y, Chen X. Anthocyanin biosynthesis in pears is regulated by a R2R3-MYB transcription factor PyMYB10. Planta. 2010;232:245–55.
**e XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012;35:1884–97.
Zhang Y, Liu Z, Liu R, Hao H, Bi Y. Gibberellins negatively regulate low temperature-induced anthocyanin accumulation in a HY5/HYH-dependent manner. Plant Signal Behav. 2011;6:632–4.
Zhang C, Jia H, Wu W, Wang X, Fang J, Wang C. Functional conservation analysis and expression modes of grape anthocyanin synthesis genes responsive to low temperature stress. Gene. 2015;574:168–77.
Zhu YC, Zhang B, Allan AC, Lin-Wang K, Zhao Y, Wang K, et al. DNA demethylation is involved in the regulation of temperature-dependent anthocyanin accumulation in peach. Plant J. 2020;102:965–76.
Wang R, Wang L, Yuan S, Li Q, Pan H, Cao J, et al. Compositional modifications of bioactive compounds and changes in the edible quality and antioxidant activity of 'Friar' plum fruit during flesh reddening at intermediate temperatures. Food Chem. 2018;254:26–35.
Wang L, Sang W, Xu R, Cao J. Alteration of flesh color and enhancement of bioactive substances via the stimulation of anthocyanin biosynthesis in 'Friar' plum fruit by low temperature and the removal. Food Chem. 2020;310:125862.
Song T, Li K, Wu T, Wang Y, Zhang X, Xu X, et al. Identification of new regulators through transcriptome analysis that regulate anthocyanin biosynthesis in apple leaves at low temperatures. PLoS One. 2019;14:e0210672.
Tian J, Han ZY, Zhang LR, Song TT, Zhang J, Li JY, et al. Induction of anthocyanin accumulation in crabapple (Malus cv.) leaves by low temperatures. HortScience. 2015;50:640–9.
Azuma A, Yakushiji H, Koshita Y, Kobayashi S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta. 2012;236:1067–80.
Fang Z, Lin-Wang K, Jiang C, Zhou D, Lin Y, Pan S, et al. Postharvest temperature and light treatments induce anthocyanin accumulation in peel of ‘Akihime’ plum (Prunus salicina Lindl.) via transcription factor PsMYB10.1. Postharvest Biol Technol. 2021;179:1–11.
Li B, **a Y, Wang Y, Qin G, Tian S. Characterization of genes encoding key enzymes involved in anthocyanin metabolism of kiwifruit during storage period. Front Plant Sci. 2017;8:1–8.
Sicilia A, Scialo E, Puglisi I, Lo Piero AR. Anthocyanin biosynthesis and DNA methylation dynamics in sweet orange fruit [Citrus sinensis L. (Osbeck)] under cold stress. J Agric Food Chem. 2020;68:7024–31.
Yu M, Man Y, Wang Y. Light- and temperature-induced expression of an R2R3-MYB gene regulates anthocyanin biosynthesis in red-fleshed kiwifruit. Int J Mol Sci. 2019;20:1–22.
Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–49.
Alappat B, Alappat J. Anthocyanin pigments: beyond aesthetics. Molecules. 2020;25:1–17.
Ban Y, Honda C, Hatsuyama Y, Igarashi M, Bessho H, Moriguchi T, et al. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007;48:958–70.
Finkelstein RR, Gibson SI. ABA and sugar interactions regulating development: cross-talk or voices in a crowd? Curr Opin Plant Biol. 2002;5:26–32.
Ban Y, Kondo S, Ubi BE, Honda C, Bessho H, Moriguchi T. UDP-sugar biosynthetic pathway: contribution to cyanidin 3-galactoside biosynthesis in apple skin. Planta. 2009;230:871–81.
Hu DG, Sun CH, Zhang QY, An JP, You CX, Hao YJ. Glucose sensor MdHXK1 phosphorylates and stabilizes MdbHLH3 to promote anthocyanin biosynthesis in apple. PLoS Genet. 2016;12:1–27.
Zheng Y, Tian L, Liu H, Pan Q, Zhan J, Huang W. Sugars induce anthocyanin accumulation and flavanone 3-hydroxylase expression in grape berries. Plant Growth Regul. 2009;58:251–60.
Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39:366–80.
Petroni K, Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–29.
Naing AH, Kim CK. Roles of R2R3-MYB transcription factors in transcriptional regulation of anthocyanin biosynthesis in horticultural plants. Plant Mol Biol. 2018;98:1–18.
He Q, Ren Y, Zhao W, Li R, Zhang L. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of purple head chinese cabbage (Brassica rapa L.). Genes (Basel). 2020;11:1–23.
Palapol Y, Ketsa S, Lin-Wang K, Ferguson IB, Allan AC. A MYB transcription factor regulates anthocyanin biosynthesis in mangosteen (Garcinia mangostana L.) fruit during ripening. Planta. 2009;229:1323–34.
Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, et al. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010;10:1–17.
Ravaglia D, Espley RV, Henry-Kirk RA, Andreotti C, Ziosi V, Hellens RP, et al. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013;13:1–14.
Medina-Puche L, Cumplido-Laso G, Amil-Ruiz F, Hoffmann T, Ring L, Rodriguez-Franco A, et al. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria x ananassa fruits. J Exp Bot. 2014;65:401–17.
Starkevic P, Paukstyte J, Kazanaviciute V, Denkovskiene E, Stanys V, Bendokas V, et al. Expression and anthocyanin biosynthesis-modulating potential of sweet cherry (Prunus avium L.) MYB10 and bHLH genes. PLoS One. 2015;10:e0126991.
** W, Feng J, Liu Y, Zhang S, Zhao G. The R2R3-MYB transcription factor PaMYB10 is involved in anthocyanin biosynthesis in apricots and determines red blushed skin. BMC Plant Biol. 2019;19:287.
Rahim MA, Busatto N, Trainotti L. Regulation of anthocyanin biosynthesis in peach fruits. Planta. 2014;240:913–29.
Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007;49:414–27.
Wang N, Qu C, Jiang S, Chen Z, Xu H, Fang H, et al. The proanthocyanidin-specific transcription factor MdMYBPA1 initiates anthocyanin synthesis under low-temperature conditions in red-fleshed apples. Plant J. 2018;96:39–55.
Todd CD, Zeng P, Huete AM, Hoyos ME, Polacco JC. Transcripts of MYB-like genes respond to phosphorous and nitrogen deprivation in Arabidopsis. Planta. 2004;219:1003–9.
Abe M, Kaya H, Watanabe-Taneda A, Shibuta M, Yamaguchi A, Sakamoto T, et al. FE, a phloem-specific Myb-related protein, promotes flowering through transcriptional activation of FLOWERING LOCUS T and FLOWERING LOCUS T INTERACTING PROTEIN 1. Plant J. 2015;83:1059–68.
Zhang Y, Mitsuda N, Yoshizumi T, Horii Y, Oshima Y, Ohme-Takagi M, et al. Two types of bHLH transcription factor determine the competence of the pericycle for lateral root initiation. Nat Plants. 2021;7:633–43.
Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–80.
Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. PNAS. 2004;101:4706–11.
An JP, Wang XF, Li YY, Song LQ, Zhao LL, You CX, et al. EIN3-LIKE1, MYB1, and ETHYLENE RESPONSE FACTOR3 act in a regulatory loop that synergistically modulates ethylene biosynthesis and anthocyanin accumulation. Plant Physiol. 2018;178:808–23.
Zhang J, Xu H, Wang N, Jiang S, Fang H, Zhang Z, et al. The ethylene response factor MdERF1B regulates anthocyanin and proanthocyanidin biosynthesis in apple. Plant Mol Biol. 2018;98:205–18.
Nowak K, Gaj MD. Stress-related function of bHLH109 in somatic embryo induction in Arabidopsis. J Plant Physiol. 2016;193:119–26.
Wang M, Jiang N, Wang Y, Jiang D, Feng X. Characterization of phenolic compounds from early and late ripening sweet cherries and their antioxidant and antifungal activities. J Agric Food Chem. 2017;65:5413–20.
Gao Y, Wang M, Jiang N, Wang Y, Feng X. Use of ultra-performance liquid chromatography-tandem mass spectrometry on sweet cherries to determine phenolic compounds in peel and flesh. J Sci Food Agric. 2019;99:3555–62.
Zhebentyayeva T, Shankar V, Scorza R, Callahan A, Ravelonandro M, Castro S, et al. Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genoty**. Hortic Res. 2019;6:1–13.
Gasic K, Hernandez A, Korban SS. RNA extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Report. 2004;22:437–8.
Acknowledgements
We would like to acknowledge all the teachers and students from the laboratory of the College of Life Science of Hebei Normal University and the Agriculture Institute of Biotechnology and Food Science of Hebei Academy of Agriculture and Forestry Sciences for their help.
Funding
This work was supported by the HAAFS Agriculture Science and Technology Innovation Project (2019-2-1), the National Natural Science Foundation of China (315018012) and the Young Talents Fund of Hebei province.
Author information
Authors and Affiliations
Contributions
J.F.G and Y.D.C conceived the project and designed the experiments. X.L.L performed the experiment and write the manuscript. M.W performed the phenolic acid and flavonoid detection. S.J.C, J.F.G and Y.D.C guided the operation of the experiment and assisted with writing. The author(s) read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Figure S1
. GO enrichment of the genes in turquoise module. Figure S2. KEGG enrichment of the genes in turquoise module. Figure S3. Correlation analysis of expression profiles of anthocyanin biosynthesis related genes, anthocyanin components and quercetin content. Table S1. The primers used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, X., Cheng, Y., Wang, M. et al. Weighted gene coexpression correlation network analysis reveals a potential molecular regulatory mechanism of anthocyanin accumulation under different storage temperatures in ‘Friar’ plum. BMC Plant Biol 21, 576 (2021). https://doi.org/10.1186/s12870-021-03354-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-021-03354-2