Abstract
Background
Gnetum is an economically important tropical and subtropical gymnosperm genus with various dietary, industrial and medicinal uses. Many carbohydrates, proteins and fibers accumulate during the ripening of Gnetum seeds. However, the molecular mechanisms related to this process remain unknown.
Results
We therefore assembled a full-length transcriptome from immature and mature G. luofuense seeds using PacBio sequencing reads. We identified a total of 5726 novel genes, 9061 alternative splicing events, 3551 lncRNAs, 2160 transcription factors, and we found that 8512 genes possessed at least one poly(A) site. In addition, gene expression comparisons of six transcriptomes generated by Illumina sequencing showed that 14,323 genes were differentially expressed from an immature stage to a mature stage with 7891 genes upregulated and 6432 genes downregulated. The expression of 14 differentially expressed transcription factors from the MADS-box, Aux/IAA and bHLH families was validated by qRT-PCR, suggesting that they may have important roles in seed ripening of G. luofuense.
Conclusions
These findings provide a valuable molecular resource for understanding seed development of gymnosperms.
Similar content being viewed by others
Background
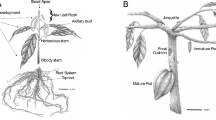
Gnetum is a genus of tropical and subtropical gymnosperm trees and shrubs distributed in South America, eastern Africa, and Asia [1]. Gnetum possesses remarkable economic potential for dietary and industrial use: its leaves are used as a vegetable, its stems and bark are made into string, nets and paper, and its seeds are used in oil and drinks. A Gnetum seed originates from a female reproductive unit that is produced on the collar involucre of a female strobilus [1, 2]. A Gnetum seed is composed of three layers of envelopes, the outermost of which gives rise to a seed coat—aril [3, 4]. Gnetum seeds are rich in a variety of chemicals, such as carbohydrates, proteins and fibers [5, 6]. The primary metabolism (e.g. carbohydrate metabolism) is probably associated with seed ripening process (during which the aril color changes from green to red, Fig. 1) in Gnetum, but the molecular mechanisms that underlie the process have not been carefully investigated.
Previous investigations of transcription factors (TFs) provide valuable insight into the molecular mechanisms of reproductive organ development in Gnetum. MADS-box genes, comprising type I and type II MADS-box genes, encode essential transcription factors that regulate reproductive organ development in seed plants [7]. Previous work has shown that type II MADS-box AG-like and TM8-like genes are highly expressed in G. luofuense seeds [8]. In addition, Aux/IAA genes participate in the development of various organs in seed plants by responding to the hormone auxin/indole-3-acetic acid [9, 10]. A recent study showed that six Aux/IAA genes are involved in the development of female strobili in G. luofuense [11]. Another study reported that bHLH genes facilitate the development of G. luofuense leaves [11]. Moreover, bHLH and MYB TFs are able to form a complex that regulates stamen development and seed production [1D); of these, 63,049 uniquely mapped reads (38.01% of total mapped reads) were mapped to the positive strand of the reference genome, 60,292 uniquely mapped (36.35%) reads were mapped to the negative strand, 39,546 (23.84%) were multiply mapped reads, and 2996 (1.81%) reads were unmapped. The map** density on each scaffold of G. luofuense genome was shown in Fig. S1E. Over 98% of the mapped reads showed similarity to the reference genome, and coverage values of the mapped reads were all above 80% (Fig. S1F). After deleting the unmapped and redundant reads, 41,151 reads remained, of which 7899 were novel isoforms of known genes and 5726 reads were from novel genes.
Annotation and classification of novel genes
The 5726 novel genes were annotated by searching against six databases—NCBI NR, KEGG, GO, SwissProt, KOG, and Pfam. A total of 4099 novel genes were annotated, of which 2588 were annotated in the NR database (Table S3). Five species—Picea sitchensis (649 genes), Amborella trichopoda (116), Vitis vinifera (88), Elaeis guineensis (80), and Nelumbo nucifera (61)—produced the largest numbers of hits to the G. luofuense novel genes (Fig. S2A). Two thousand four hundred eighty-seven novel genes were annotated with KEGG pathways (Table S3), and the most enriched pathways were “signal transduction” (169 genes), “carbohydrate metabolism” (83 genes), and “translation” (69 genes, Fig. S2B). GO analysis classified 2069 genes into three categories: “biological process”, “cellular components” and “molecular functions” (Fig. S2C). Novel genes classified in the biological process category were mainly annotated with the terms “metabolic process” (1052), “cellular process” (1037), and “single-organism process” (581). Novel genes classified in the cellular component category were mainly annotated with the terms “cell” (519), “cell part” (519), and “membrane” (367). Novel genes classified in the molecular function category were mainly annotated with the terms “binding” (1192), “catalytic activity” (942), and “transporter activity” (132). One thousand nine hundred thirty genes, 1315 genes and 2069 genes were annotated with the Swiss Prot, KOG and Pfam databases, respectively (Table S3).
AS and APA analysis
After map** reads to the reference genome of G. luofuense, a total of 9061 AS events were detected. These could be classified into seven types (Fig. 2a): retained intron (2713, 29.94%), alternative 3′ splice site (2468, 27.24%), alternative 5′ splice site (1769, 19.52%), skipped exon (1305, 14.40%), alternative first exon (542, 5.98%), alternative last exon (217, 2.39%), and mutually exclusive exon (47, 0.52%).
a Numbers of alternative splicing events identified in the full-length transcriptome of G. luofuense. b PCR validation of AS events, i.e. retained intention (at top) and skipped exon (at bottom) of two selected genes. c Genes with different numbers of alternative polyadenylation sites identified in the full-length transcriptome of G. luofuense seeds
To verify the AS events identified, expression of two genes, i.e. Tns00138667g03 and TnS000973269g04 were validated by qRT-PCR (Fig. 2b, Additional file 1). In addition, a total of 8512 genes from G. luofuense seeds had at least one supported poly(A) site. Of these, 3654 (42.93%) had a single poly(A) site, and 640 (7.52%) had at least five poly(A) sites (Fig. 2c). The largest number of poly(A) sites—21—was found in the gene TnS000670009g01.
Identification of TFs and lncRNAs
A total of 2160 transcription factors (TFs) from 86 gene families were detected using iTAK. The largest fraction of identified TFs came from the C3H (5.6%), bHLH (4.53%), and MYB-related (4.26%) families (Fig. 3a). In addition, 11,885, 5958, 11,294 and 11,037 lncRNAs were identified using the CNCI, CPC, PFAM and PLEK methods, respectively. A total of 3551 lncRNAs were identified by all four methods (Fig. 3b), with lengths ranging from 200 to 7840 bp. The lncRNAs were further classified into four types (Fig. 3c): 1422 (40.05%) sense intronic lncRNA, 1149 (32.36%) long intergenic non-coding RNA, 547 (15.40%) antisense lncRNA, and 433 sense overlap** lncRNA (12.19%). The length distribution of the identified lncRNAs was considerably narrower than that of mRNAs predicted from the G. luofuense genome (Fig. 3d). Moreover, most identified lncRNAs had five or fewer exons, whereas mRNAs predicted from the reference genome tended to have larger numbers of exons (Fig. 3e).
a A partial list of transcription factors (top 28 gene families) identified in the full-length transcriptome of G. luofuense seeds. b Venn diagram showing the number of lncRNAs identified using four different approaches: CPC (Coding Potential Calculator), CNCI (Coding-Non-Coding Index), CPAT (Coding Potential Assessment Tool), and Pfam (Protein Family). c Functional classification and numbers of four lncRNA types. d The length density distribution of identified lncRNAs on the reference genome of G. luofuense compared to that of identified lncRNA in the full-length transcriptome. e Distribution of exon numbers in mRNAs predicted by the reference genome and identified lncRNAs in the full-length transcriptome
Illumina sequencing of seed samples at two developmental stages
To explore gene expression patterns during seed development of G. luofuense, 306,900,384 clean Illumina reads (46.04 Gb of raw data) with Q30 values from 93.54 to 94.07% were generated from three immature seed samples (IS) and three mature seed samples (MS) (Table S4). After the deletion of adaptors and low-quality reads, the average GC content of the six samples was 47.08%. PCA analysis showed that gene expression was highly correlated among the replicate samples of immature and mature seeds (correlation efficiency value = 0.95, cumulative proportion of variation explained by PC1 and PC2 = 78.7%) (Fig. 4a). After map** to the G. luofuense genome, the map** ratios of IS (average 89.44%, Table S5) were found to be significantly larger than those of MS (average 84.46%, Student’s t-test p-value = 0.003). RNA-seq analysis of the two developmental stages yielded a total of 23,977 genes (19,010 in IS and 20,737 in MS), of which 2970 were identified as novel genes.
Detection of DEGs and qRT-PCR validation. a PCA analysis of gene expression in the three immature seed samples (IS01–03) and three mature seed samples (MS01–03). b A volcano plot of differential gene expression between immature and mature seed samples of G. luofuense, with upregulated genes in red and downregulated genes in green from immature seeds to mature seeds. c A bubble plot of enriched GO terms; the x-axis represents the z-score, the y-axis represents the negative logarithm of the adjusted p-values, the circle sizes are proportional to the number of genes enriched in the GO terms, and the circle colors denote the three GO term categories. A table on right side describes top 14 GO terms in the category biological process, which have been labeled in the bubble plot on left side. d A bubble plot of enriched KEGG terms; the x-axis represents rich factors, the circle sizes are proportional to enriched gene numbers, and the circle colors correspond to the negative logarithm of the adjusted p-values for each KEGG pathway. e The expression of 14 TF genes (i.e. MADS-box, Aux/IAA, bHLH and MYB genes) from immature and mature seeds of G. luofuense were verified by qRT-PCR, and the expression values were normalized with the △△Ct-method
Enrichment analysis of DEGs and qRT-PCR validation
A total of 14,323 differentially expressed genes (DEGs) were identified between IS (control group) and MS: we found 7891 upregulated genes and 6432 genes downregulated (Fig. 4b) from IS to MS. The DEGs were also annotated with the three categories of GO terms, and multiple GO terms in the “biological process” category were significantly enriched with regard to Z-scores and adjusted p-values (Fig. 4c). The top five enriched GO terms were “single-organism cellular process” (GO:0044763), “single-organism process” (GO:0044699), “metabolic process” (GO:0008152), “cellular metabolic process” (GO:0044237), and “Organic substance metabolic process” (GO:0071704). The DEGs were also enriched in multiple KEGG pathways with reference to Arabidopsis thaliana. The top five enriched KEGG pathways were “metabolic pathways” (KEGG ID: ath01100, 1229 genes), “biosynthesis of secondary metabolites” (ath01110, 844 genes), “carbon metabolism” (ath01200, 179 genes), “ribosome” (ath03010, 164 genes), and “starch and sucrose metabolism” (ath00500, 154 genes) (Fig. 4d). qRT-PCR was used to validate the relative expression of 14 genes of interest: four MADS-box genes, four Aux/IAA genes, four bHLH genes, and two MYB genes. The relative expression of the 14 genes at the two seed developmental stages is presented in Fig. 4e.
Discussion
Structural analysis of the full-length transcriptome
Structural analysis of the full-length transcriptome
AS event analysis
In angiosperms, it has been reported that the percentages of AS modes differ dramatically among organs and that vegetative tissues (e.g. roots and ears) exhibit higher percentages of intron retention than reproductive tissues (e.g. pollen and endosperm) in maize (see Fig. 3 in 17). Moreover, retained intron percentage dramatically declines over the course of fruit development in strawberry [30]. In gymnosperms, Gingko biloba genes (e.g. GB_12621 and GB_20198) show differences in AS between vegetative and reproductive organs and between immature and mature stages of leaves and seeds [26]. In the present study, retained intron accounted for 29.94% of all AS events (Fig. 2a), this figure was dramatically lower than those reported previously in leaves [41.5%, 12] and female strobili [46%, 11] of G. luofuense. This result suggests that the frequency of different AS modes may not only be species specific but may also vary among different organs.
APA analysis
In angiosperms, alternative polyadenylation is involved in the regulation of flowering time [31, 32]. In gymnosperms, it has been suggested that investigation of APA can improve genome annotation and promote understanding of flavonoid biosynthesis in G. biloba [26]. In the present study, a total of 8512 genes were identified as having at least one poly(A) site, the numbers of genes with various numbers of poly(A) sites declined dramatically as the number of poly(A) sites increased from one to five (Fig. 2c). The pattern of gene numbers with various numbers of poly(A) sites is consistent with that observed in G. luofuense leaves and female strobili [11]. Our results suggest that alternative polyadenylation enriches the proteomic complexity and affects the seed ripening process of G. luofuense.
lncRNA analysis
In angiosperms, lncRNAs participate in fruit development and color change in strawberry [33], as well as aroma formation in black tea [34]. In gymnosperms, lncRNAs have an important role in the regulation of leaf development [27] and leaf color changes in G. biloba [25]. The percentages of four lncRNA types have been shown to differ dramatically in G. biloba: lincRNA (50.6%), sense lncRNA (21.6%), intronic lncRNA (20.9%), and antisense lncRNA (6.9%) [26]. A recent study show that lincRNA was the highest (40.8%) and antisense lncRNA was the lowest (1.67%) in the full-length transcriptome of the G. luofuense female strobilus [11]. In the present study, however, we found number of intronic lncRNA (40.05%) was the highest, and that of sense lncRNA (2.1%) was the lowest (Fig. 3c). Thus, it appears that G. luofuense uses different lncRNAs to regulate the different reproductive organs. The scenario is similar to the results reported in P. abies [35]. Moreover, lncRNAs tended to be shorter and possessed fewer exons than protein coding genes [27], this finding is consistent with previous studies in gymnosperms, such as G. biloba [27], Picea abies [35], and G. luofuense [11].
Key TFs/genes involved in seed ripening of G. luofuense
MADS-box genes
MADS-box transcription factors are classified into type I and type II groups based on the sequence of the conserved MADS domain [36, 37]. Compared with type II genes, type I genes have received less attention in previous studies, although their roles in the development of female gametophytes, embryos and seeds have been highlighted in angiosperms [36, 38]. The functions of type I genes are poorly understood in gymnosperms, and broad expression of type I genes in shoots, needles and strobili of conifers has been regarded as “transcriptional noise” [39]. Type I genes are further divided into Mα, Mβ and Mδ subgroups; Mα genes are generally expressed in various shoot tissues of conifers, whereas Mβ/Mδ genes are expressed in embryos, buds, and male strobili [39]. In G. luofuense, a total of 11 type I genes, (seven Mα genes, three Mβ genes, and one Mδ gene) have been reported [8]. Twenty-seven type I MADS-box TFs were identified in G. luofuense seeds (Fig. 4e), and gene TnS000803113g11 was differentially expressed between immature and mature seeds, indicating an important role in seed ripening of G. luofuense.
Among type II MADS-box genes, the expression of TM8 genes was first reported in tomato flowers [40] and the TM8-like gene ERAF17 was shown to be expressed in female flowers but not male flowers of cucumber [41]. In gymnosperms, TM8-like genes (e.g. GbMADS11 and GbMADS6 in G. biloba and TbTM8 in Taxus baccata) influence aril development in male strobili and seed aril develo** [42]. In G. luofuense, TM8-like genes accounted for almost half the number of genes with a varied pattern of expression [8]. For example, TnS013912549g01 was expressed in both female and male strobili, whereas TnS001008199t01 was exclusively expressed in male strobili [8]. A previous study showed that GpMADS1, a TM8-like gene defined in Hou et al. (2019b), participated in the development of female strobili in G. parvifolium [43]. Thus far, 38 type II genes have been identified in G. luofuense, of which TM8-like genes constitute almost half of the identified gene numbers [8]. In the present study, two TM8-like genes TnS000061251g01 and TnS000980857g01 were differentially expressed. Moreover, other type II MADS-box genes, such as AG-like gene TnS000064931g01 and AGL6-like gene TnS000229425g02 were differentially expressed at two developmental stages of G. luofuense seeds. Our results are congruent with a previous study that AG, AGL6 and TM8-like genes regulate seed development of G. biloba and T. baccata [42].
Aux/IAA genes
Aux/IAA TFs play an essential role in the auxin responses of seed plants [9, 10, 44]. For example, in angiosperms, FaAux/IAA1 and Aux/IAA2 participate in the fruit development of strawberry [45], and EgrIAA4 is thought to be essential to the regulation of secondary cell wall and fiber development in Eucalyptus [46]. Another study showed that IAA9 was involved in fruit and leaf morphogenesis in tomato [47] In gymnosperms, the Aux/IAA gene LaIAA2 appears to be important for the regulation of root development and auxin signaling [48]. Besides, six Aux/IAA genes (GluIAA1–6) have been identified in G. luofuense, all of which are involved in female strobilus development [11]. In the present study, four Aux/IAA genes, TnS000653177g04 (GluIAA2), TnS000867017g28 (GluIAA3), TnS000053353g02 (GluIAA4), and TnS000142615g19 (GluIAA5), were differntially expressed and validated by qRT-PCR between the two developmental stages of G. luofuense seeds (Fig. 4e). These results suggest that Aux/IAA genes may also be of importance in G. luofuense seed ripening.
bHLH genes
In angiosperms, the bHLH-encoding gene SPATULA has been reported to control the development of flowers and fruits in Arabidopsis [49, 50], and a bHLH TF has been shown to determine seed coat color in Brassica rapa [51]. Moreover, bHLH TFs, together with MYB and WDR TFs, are involved in the regulation of flavonoid biosynthesis [52,53,54]. The expression of two MYB-related genes, i.e. Osmyb1 and Osmyb4, reaches the level of saturation at 14 days after the anthesis, suggesting that they have an important role in the maturation of rice seeds [55]. In gymnosperms, three bHLH TFs have been reported to negatively regulate gene expression in the paclitaxel biosynthesis pathway in response to jasmonate in Taxus cuspidata [56]. Besides, bHLH and MYB TFs have been reported to participate in flavonoid biosynthesis in the roots rather than the seeds of Ginkgo biloba [26]. In G. luofuense, 67 bHLH genes were identified in leaves based on full-length transcripts; 30 were subjected to phylogenetic analysis and classified into four subgroups [57]. Furthermore, 110 bHLH TFs were the most abundant TFs during development of the female strobilus in G. luofuense [11]. In the present study, 98 bHLH were identified, of which four bHLH genes, i.e. Tns000226135g02, TnS000896885g01, TnS000889809g02, and TnS000498063g28 were differentially expressed and their expression was validated by qRT-PCR (Fig. 4e). These results suggest that bHLH TFs may also play an imporant role in seed ripening of G. luofuense.
Genes related to carbohydrate metabolism
Gnetum seeds are rich in carbohydrates as the examples reported in G. africanum (87.62%) [5] and G. gnemon (64.1%) [6]. The accumulation of carbohydrates in Gnetum seeds makes them palatable and nutritious, thereby attracting a variety of herbivores to promote seed dispersal [58, 59]. In the present study, the DEGs between immature and mature seeds were enriched in several KEGG pathways, e.g. carbon metabolism, starch and sucrose metabolism, glycolysis/gluconeogenesis, and fructose and mannose metabolism (Fig. 4d). The DEGs were also enriched in the GO terms, e.g. primary metabolic process, metabolic process, and cellular metabolic process (Fig. 4c). These results suggest that genes that are involved in carbohydrate metabolisms are also indispensable in seed ripening of G. luofuense.
Conclusions
We generated a full-length transcriptome of G. luofuense seeds at two developmental stages using Pacbio sequencing technique. We identified a total of 5726 novel genes, 9061 alternative splicing events, 3551 lncRNAs, and 8512 genes were identified to possess at least one poly(A) site. Transcription factors MADS-box, Aux/IAA and bHLH were found to play important roles in seed ripening of G. luofuense. These findings provide a valuable molecular resource for disentangling organ development of gymnosperms.
Methods
Plant material and RNA extraction
Gnetum luofuense seeds were collected at immature (IS) and mature (MS) developmental stages from a female individual (voucher number “CH003”, SYS) cultivated in the Bamboo Garden at Sun Yat-sen University on September 2nd and 28th 2018 (Fig. 1a) with the permissions of Sun Yat-sen University. To obtain a full-length transcriptome for the two developmental stages, identical amounts (10 g) of mature and immature seeds with arils were pooled, incubated in liquid nitrogen, and frozen at − 20 °C for PacBio SMRT sequencing. In addition, six samples of G. luofuense seeds (“IS001–003” and “MS001–003”) were collected for Illumina sequencing, three from the immature stage (control group) and three from the mature stage. The RNA for each sample was extracted using an RNA kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. RNase-free DNase (Qiagen) was used to remove relic DNA, and the RNA concentration of samples was evaluated by 1% agarose gel electrophoresis. A NanoDrop spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) were used to assess the purity and integrity of the extracted RNA. G. luofuense samples used in this research is derived from the plant cultivated merely for teaching and researches. Thus, the collection of seeds and the performance of experimental research on such plant were complied with the national guidelines of China.
Plant material and RNA extraction
Gnetum luofuense seeds were collected at immature (IS) and mature (MS) developmental stages from a female individual (voucher number “CH003”, SYS) cultivated in the Bamboo Garden at Sun Yat-sen University on September 2nd and 28th 2018 (Fig. 1a). To obtain a full-length transcriptome for the two developmental stages, identical amounts (15 g) of mature and immature seeds with arils were pooled, incubated in liquid nitrogen, and frozen at − 20 °C for PacBio SMRT sequencing. In addition, the six samples of G. luofuense seeds were collected for Illumina sequencing, three from the immature stage (control group) and three from the mature stage. The RNA for each sample was extracted using an RNA kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions. RNase-free DNase (Qiagen) was used to remove relic DNA, and the RNA concentration of samples was evaluated by 1% agarose gel electrophoresis. A NanoDrop spectrophotometer (ThermoFisher Scientific, Wilmington, DE, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) were used to assess the purity and integrity of the extracted RNA.
Library construction and PacBio sequel sequencing
When the integrity of extracted RNA met the minimum requirement (> 7.0), full-length cDNA was synthesized using a SMARTer PCR cDNA Synthesis kit (Clontech, Takara Bio Inc., Shiga, Japan). The synthesized cDNA was subjected to PCR amplification using a KAPA HIFI PCR kit (Kapa Biosystems, Boston, MA, USA). After PCR amplification, the cDNA was quality controlled and purified using a QIAquick PCR Purification kit (Qiagen, Hilden, Germany). The RNA samples were subjected to terminal repair and the attachment of SMRT dumbbell-type adapters. Before PacBio sequencing, two bins (1–4 kb, 4–6 kb) were established to preferentially sequence the smaller cDNAs.
Library construction and Illumina sequencing
Before Illumina sequencing, all six RNA samples that possessed poly(A) were enriched with oligo (dT) magnetic beads. The enriched RNA was randomly reduced to small pieces with a fragmentation buffer. First strand cDNA was generated using hexamers and reverse transcriptase (Superscript III, Invitrogen). After purification with AMPure XP beads, second strand cDNA was synthesized using DNA polymerase I, RNase H and dNTPs (Sigma-Aldrich). The double-stranded cDNA was subjected to terminal repair and poly(A) tailing, followed by Illumina adaptor ligation. The final cDNA library was completed after a second round of purification and PCR amplification. The quality of the six cDNA libraries was assessed using a Qubit 2.0 fluorometer prior to sequencing on the Illumina HiSeq 4000 platform.
PacBio data processing and error correction
PacBio sequencing data were analyzed using PacBio SMRTlink v. 5.1 software. First, we obtained reads of inserts (ROIs) from the BAM files generated from the platform using the following parameters: maximum drop fraction—0.8, minimum length—200, no polish, minimum z-score—9999, minimum passes—1, minimum predicted accuracy—0.8, and maximum length—18,000. The ROIs were classified into full-length reads (FLs) and non-full-length reads (nFLs) based on the presence and absence of 5′ and 3′ cDNA primers and a 3′ poly (A) tail, see also in [11]. The FLs and nFLs were clustered to achieve consensus isoforms using an isoform-level clustering (ICE) algorithm. To obtain full-length non-chimeric (FLNC) isoforms, the high-quality isoforms from FLs were corrected using Quiver software with a post-correction accuracy above 99%. The low-quality consensus isoforms from nFLs were further corrected with LoRDEC [60] using two Illumina-sequenced samples (one from mature seeds and one from immature seeds).
Genome map** and novel gene detection
All FLNCs and corrected nFLs were mapped to the reference genome of G. luofuense (=G. montanum) [61]. The GMAP output files were used for subsequent analyses. Redundant FLNCs were removed using the following parameters: minimum identity—0.9, minimum trimmed coverage—0.85, and allow close indel—0. Mapped FLNCs with different lengths at their 5′ ends were not considered to be redundant. The FLNCs that mapped to annotated genes in the G. luofuense genome were considered to be known genes; otherwise, they were classified as novel genes and novel isoforms of known genes.
Functional annotation and classification
All identified novel genes were annotated by BLASTX v.2.2.26 searches (E-value < 1 × 10− 5) of the gene ontology (GO, http://www.geneontology.org), Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/), Protein Family (Pfam), KOG/COG (Clusters of Orthologous Groups of proteins, http://www.ncbi.nlm.nih.gov/COG/), NCBI non-redundant protein sequence (NR, http//www.ncbi.nlm.nih.gov/), and Swiss-Prot (http// http://www.expasy.org/sprot/) databases and by HMMER v.3.1b2 searches (E-value < 1 × 10− 10) of the Pfam (Protein Family, http://pfam.xfam.org/) database [62, 63]. In addition, GO enrichment analysis was performed using the GOseq package implemented in R [64] and KEGG enrichment analysis was performed using KOBAS version 2.0 [65].
AS and APA analysis
Gene structure analysis was performed using the TAPIS pipeline [16]. First, seven types of alternative splicing (AS) events were identified: alternative 3′ splice site, retained introns, alternative 5′ splice site, skipped exon, alternative first exon, alternative last exon, and mutually exclusive exons. In addition, alternative polyadenylation (APA) analysis was conducted, and genes were classified according to their poly(A) number.
Identification of TFs and lncRNAs
Coding sequences (CDS), which possess open reading frames (ORFs), were identified by searching against the Pfam database using TransDecoder [66]. Based on the identified CDS, transcription factors (TFs) were predicted by searching against the Plant Transcription Factor Database v.4.0 (http://planttfdb.cbi.pku.edu.cn) using iTAK version 15.03 [67]. Four methods were used to identify lncRNAs: PC (Coding Potential Calculator), CNCI (Coding-Non-Coding Index), CPAT (Coding Potential Assessment Tool), and Pfam. The lncRNAs, which are longer than 200 nt and possess at least two exons, do not encode proteins and are classified into four groups: lincRNA, intronic lncRNA, sense lncRNA, and antisense lncRNA.
DEG identification and qRT-PCR validation
Illumina sequenced raw reads with poly(N) and low scores were removed, and the remaining reads were trimmed of adaptors at both ends. The cleaned reads were mapped to the G. luofuense genome using HISAT2 v.2.1.0 [68]. Mapped read numbers were counted and adjusted through one scaling normalized factor using the R package edgeR [69]. The numbers of mapped reads were converted to values of fragments per kilobase of transcript per million mapped fragments (FPKM). To identify differentially expressed genes (DEGs), RNA data from three replicate samples of mature and immature seeds were separately merged and then compared using the R package EBSeq v. 1.20.0 [70]. The DEGs met the following requirements: corrected P-value (adjusted by the Benjamini & Hochberg method)—0.005 and log2(fold change) value—1.
qRT-PCR validation
To validate the occurrence of AS events identified in the full-length transcriptome, expression of two genes, i.e. TnS000138667g03 and TnS000973269g04 were validated by qRT-PCR. Among the two micrograms of RNA were extracted from mature and immature seeds of G. luofuense and subjected to cDNA synthesis according to the manufacturer’s protocol. qRT-PCR was performed under the following conditions: 10 min at 95 °C (1 cycle), 10 s at 95 °C, 30 s at 55 °C and 15 s at 72 °C (40 cycles), temperature reduction from 95 °C to 60 °C (0.5 °C/10 s) and termination in 30 s at 25 °C. Gene electrophorese was performed to test various lengths of qRT-PCR products. Moreover, 14 DEGs were selected for gene expression validation with qRT-PCR. The G. luofuense actin gene was used as an endogenous control to estimate the relative expression of target genes using the ΔΔCt-method [71]. For each sample, three replicates were performed, and the mean and standard deviation of the qRT-PCR gene expression values were calculated accordingly.
Availability of data and materials
PacBio sequencing data from the merged seed sample and RNA-seq data from the six samples were all deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession number PRJNA622631. The data that support the results are included within the article and its additional files. Other relevant materials are available from the corresponding authors on reasonable request.
Abbreviations
- DEGs:
-
Differentially expressed genes
- CDS:
-
Coding sequences
- ORFs:
-
Open reading frames
- TFs:
-
Transcription factors
- PC:
-
Coding potential calculator
- CNCI:
-
Coding-non-coding index
- CPAT:
-
Coding potential assessment tool
- APA:
-
Alternative polyadenylation
- FLs:
-
Full-length reads
- nFLs:
-
Non-full-length reads
- Aux/IAA:
-
Auxin/indole-3-acetic acid
- bHLH:
-
Basic helix-loop-helix
References
Hou C, Humphreys AM, Thureborn O, Rydin C. New insights into the evolutionary history of Gnetum (Gnetales). Taxon. 2015;64(2):239–53.
Hou C, Wikström N, Strijk J, Rydin C. Resolving phylogenetic relationships and species delimitations in closely related gymnosperms using high-throughput NGS, sanger sequencing and morphology. Plant Syst Evol. 2016;302(9):1345–65.
Thoday MG. The female inflorescence and ovules of Gnetum africanum with notes on Gnetum scandens. Ann Bot. 1911;25(4):1101–35.
Berridge EM. On some points of resemblance between gnetalean and Bennettitean seeds. New Phytol. 1911;10(4):140–4.
Ekop A. Determination of chemical composition of Gnetum africanum (AFANG) seeds. Pak J Nutr. 2007;6(1):40–3.
Bhat R, Binti Yahya N. Evaluating belinjau (Gnetum gnemon L.) seed flour quality as a base for development of novel food products and food formulations. Food Chem. 2014;156:42–9.
Melzer R, Wang YQ, Theissen G. The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Semin Cell Dev Biol. 2010;21(1):118–28.
Hou C, Li L, Liu Z, Su Y, Wan T. Diversity and expression patterns of MADS-box genes in Gnetum luofuense—implications for functional diversity and evolution. Trop Plant Biol. 2020;13:36–49.
Luo J, Zhou J, Zhang J. Aux/IAA gene family in plants: molecular structure, regulation, and function. Int J Mol Sci. 2018;19(1):259.
Wu WT, Liu YX, Wang YQ, Li HM, Liu JX, Tan JX, He JD, Bai JW, Ma HL. Evolution analysis of the Aux/IAA gene family in plants shows dual origins and variable nuclear localization signals. Int J Mol Sci. 2017;18(10):107.
Hou C, Deng N, Su YJF. PacBio long-read sequencing reveals the transcriptomic complexity and Aux/IAA gene evolution in Gnetum (Gnetales). Forests. 2019;10(11):1043.
Qi T, Huang H, Song S, **e D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell. 2015;27(6):1620–33.
Yang Z, Yang Z, Yang C, Wang Z, Chen D, **e Y, Wu Y. Identification and genetic analysis of alternative splicing of long non-coding RNAs in tomato initial flowering stage. Genomics. 2020;112(1):897–907.
Wang B, Brendel V. Genome-wide comparative analysis of alternative splicing in plants. Proc Natl Acad Sci U S A. 2006;103(18):7175–80.
Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496.
Abdel-Ghany SE, Hamilton M, Jacobi JL, Ngam P, Devitt N, Schilkey F, Ben-Hur A, Reddy AS. A survey of the sorghum transcriptome using single-molecule long reads. Nat Commun. 2016;7:11706.
Wang T, Wang H, Cai D, Gao Y, Zhang H, Wang Y, Lin C, Ma L, Gu L. Comprehensive profiling of rhizome-associated alternative splicing and alternative polyadenylation in moso bamboo (Phyllostachys edulis). Plant J. 2017;91(4):684–99.
Liu XX, Mei WB, Soltis PS, Soltis DE, Barbazuk WB. Detecting alternatively spliced transcript isoforms from single-molecule long-read sequences without a reference genome. Molec Ecol Resour. 2017;17(6):1243–56.
Chao Q, Gao ZF, Zhang D, Zhao BG, Dong FQ, Fu CX, Liu LJ, Wang BC. The developmental dynamics of the Populus stem transcriptome. J Pl Biotech. 2019;17(1):206–19.
Chao Y, Yuan J, Li S, Jia S, Han L, Xu L. Analysis of transcripts and splice isoforms in red clover (Trifolium pratense L.) by single-molecule long-read sequencing. BMC Plant Biol. 2018;18(1):300.
Hu H, Yang W, Zheng Z, Niu Z, Yang Y, Wan D, Liu J, Ma T. Analysis of alternative splicing and alternative polyadenylation in Populus alba var pyramidalis by single-molecular long-read sequencing. Front Genet. 2020;11:48.
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9.
Karlik E, Ari S, Gozukirmizi N. LncRNAs: genetic and epigenetic effects in plants. Biotechnol Biotec Eq. 2019;33(1):429–39.
Liu J, Wang H, Chua NH. Long noncoding RNA transcriptome of plants. J Pl Biotech. 2015;13(3):319–28.
Wu Y, Guo J, Wang T, Cao F, Wang G. Transcriptional profiling of long noncoding RNAs associated with leaf-color mutation in Ginkgo biloba L. BMC Plant Biol. 2019;19(1):527.
Ye J, Cheng S, Zhou X, Chen Z, Kim SU, Tan J, Zheng J, Xu F, Zhang W, Liao Y. A global survey of full-length transcriptome of Ginkgo biloba reveals transcript variants involved in flavonoid biosynthesis. Ind Crop Prod. 2019;139:111547.
Wang L, **a X, Jiang H, Lu Z, Cui J, Cao F, ** B. Genome-wide identification and characterization of novel lncRNAs in Ginkgo biloba. Trees. 2018;32(5):1429–42.
Minoche AE, Dohm JC, Schneider J, Holtgrawe D, Viehover P, Montfort M, Sorensen TR, Weisshaar B, Himmelbauer H. Exploiting single-molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 2015;16(1):184.
Wan T, Liu ZM, Li LF, Leitch AR, Leitch IJ, Lohaus R, Liu ZJ, **n HP, Gong YB, Liu Y, et al. A genome for gnetophytes and early evolution of seed plants. Nat Plants. 2018;4(2):82–9.
Li Y, Dai C, Hu C, Liu Z, Kang C. Global identification of alternative splicing via comparative analysis of SMRT- and Illumina-based RNA-seq in strawberry. Plant J. 2017;90(1):164–76.
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–7.
Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113(6):777–87.
Bai L, Chen Q, Jiang L, Lin Y, Ye Y, Liu P, Wang X, Tang H. Comparative transcriptome analysis uncovers the regulatory functions of long noncoding RNAs in fruit development and color changes of Fragaria pentaphylla. Hortic Res. 2019;6(1):42.
Varshney D, Rawal HC, Dubey H, Bandyopadhyay T, Bera B, Kumar PM, Singh NK, Mondal TK. Tissue specific long non-coding RNAs are involved in aroma formation of black tea. Ind Crop Prod. 2019;133:79–89.
Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497(7451):579–84.
Gramzow L, Theissen G. A hitchhiker's guide to the MADS world of plants. Genome Biol. 2010;11(6):214.
Theissen G. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol. 2001;4(1):75–85.
Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM. The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell. 2011;23(3):865–72.
Gramzow L, Weilandt L, Theissen G. MADS goes genomic in conifers: towards determining the ancestral set of MADS-box genes in seed plants. Ann Bot. 2014;114(7):1407–29.
Pnueli L, Abu-Abeid M, Zamir D, Nacken W, Schwarz-Sommer Z, Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991;1(2):255–66.
Ando S, Sato Y, Kamachi S, Sakai S. Isolation of a MADS-box gene (ERAF17) and correlation of its expression with the induction of formation of female flowers by ethylene in cucumber plants (Cucumis sativus L.). Planta. 2001;213(6):943–52.
Lovisetto A, Guzzo F, Tadiello A, Toffali K, Favretto A, Casadoro G. Molecular analyses of MADS-box genes trace back to gymnosperms the invention of fleshy fruits. Molec Biol Evol. 2012;29(1):409–19.
Shindo S, Ito M, Ueda K, Kato M, Hasebe M. Characterization of MADS genes in the gymnosperm Gnetum parvifolium and its implication on the evolution of reproductive organs in seed plants. Evol Dev. 1999;1(3):180–90.
Guilfoyle TJ. Aux/IAA proteins and auxin signal transduction. Trends Plant Sci. 1998;3(6):205–7.
Liu D, Chen J, Lu W. Expression and regulation of the early auxin-responsive Aux/IAA genes during strawberry fruit development. Mol Biol Rep. 2011;38(2):1187–93.
Yu H, Soler M, San Clemente H, Mila I, Paiva JA, Myburg AA, Bouzayen M, Grima-Pettenati J, Cassan-Wang H. Comprehensive genome-wide analysis of the Aux/IAA gene family in Eucalyptus: evidence for the role of EgrIAA4 in wood formation. Plant Cell Physiol. 2015;56(4):700–14.
Wang H, Jones B, Li Z, Frasse P, Delalande C, Regad F, Chaabouni S, Latche A, Pech J, Bouzayen M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell. 2005;17(10):2676–92.
Feng J, Qi L, Sun X, Lin Y, Dong J, Zhang S. Isolation and characterization of an Aux/IAA gene (LaIAA2) from Larix. Afr J Biotechnol. 2012;11(47):10640–9.
Heisler MG, Atkinson A, Bylstra YH, Walsh R, Smyth DR. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development. 2001;128(7):1089–98.
Groszmann M, Paicu T, Smyth DR. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis. Plant J. 2008;55(1):40–52.
Li X, Chen L, Hong M, Zhang Y, Zu F, Wen J, Yi B, Ma C, Shen J, Tu J. A large insertion in bHLH transcription factor BrTT8 resulting in yellow seed coat in Brassica rapa. PLoS One. 2012;7(9):e44145.
Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015;20(3):176–85.
Zhang B, Chopra D, Schrader A, Hülskamp M. Evolutionary comparison of competitive protein-complex formation of MYB, bHLH, and WDR proteins in plants. J Exp Bot. 2019;70(12):3197–209.
Nemesio-Gorriz M, Blair PB, Dalman K, Hammerbacher A, Arnerup J, Stenlid J, Mukhtar SM, Elfstrand M. Identification of Norway spruce MYB-bHLH-WDR transcription factor complex members linked to regulation of the flavonoid pathway. Front Plant Sci. 2017;8:305.
Suzuki A, Suzuki T, Tanabe F, Toki S, Washida H, Wu C-Y, Takaiwa F. Cloning and expression of five myb-related genes from rice seed. Gene. 1997;198(1–2):393–8.
Lenka SK, Nims NE, Vongpaseuth K, Boshar RA, Roberts SC, Walker EL. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front Plant Sci. 2015;6:115.
Deng N, Hou C, Liu C, Li M, Bartish I, Tian Y, Chen W, Du C, Jiang Z, Shi S. Significance of photosynthetic characters in the evolution of Asian Gnetum (Gnetales). Front Plant Sci. 2019;10:39.
Corlett RT. Characteristics of vertebrate-dispersed fruits in Hong Kong. J Trop Ecol. 1996;12(6):819–33.
Forget P-M, Hammond DS, Milleron T, Thomas R. Seasonality of fruiting and food hoarding by rodents in Neotropical forests: consequences for seed dispersal and seedling recruitment. In: Forget P-M, Hammond DS, Milleron T, Thomas R, editors. Seed dispersal and Frugivory: ecology evolution and conservation. Wallingford: CAB International; 2002. p. 241–56.
Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–14.
Wu TD, Watanabe CK. GMAP: a genomic map** and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21(9):1859–75.
Finn RD, Clements J, Eddy SR. HMMER web server: interactive sequence similarity searching. Nucl Acids Res. 2011;39:29–37.
Albert VA, Barbazuk WB, Der JP, Leebens-Mack J, Ma H, Palmer JD, Rounsley S, Sankoff D, Schuster SC, Soltis DE. The Amborella genome and the evolution of flowering plants. Science. 2013;342(6165):1241089.
R Core Team. R: A language and environment for statistical computing version 3.2.0. Vienna: R Foundation for Statistical Computing; 2018. Retrieved from https://www.R-project.org.
**e C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li C-Y, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl Acids Res. 2011;39:316–22.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494.
Zheng Y, Jiao C, Sun HH, Rosli HG, Pombo MA, Zhang PF, Banf M, Dai XB, Martin GB, Giovannoni JJ, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9(12):1667–70.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Meth. 2015;12(4):357.
Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, Haag JD, Gould MN, Stewart RM, Kendziorski C. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–43.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8.
Acknowledgements
We acknowledge TopEdit LLC for the linguistic editing and proofreading during the preparation of this manuscript. We thank Dr. **** Zeng for Gnetum seed photos. We also thank three reviewers for valuable comments on the text.
Funding
This work was supported by the Special Fund for Hunan Forestry Science and Technology (XLK201812 to N.D.), Forestry Science and Technology Innovation Project of Hunan Province (HNGYL-2019-01 to N.D.), and the Natural Science Foundation of Guangdong Province, China (No. 2018A0303130163 to C.H.). The funding body had no influence over the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
ND, CH, CL and YT conceived and designed the experiments. ND and CH wrote the manuscript. CH, BH, SS and SQ performed the experiments. BH, FM and QS analyzed the data. CL and YT administrated the project. FM and SS provided assistance in software usage. BH, MF and SS collected plant samples. ND, CH, CL and YT revised the manuscript. All authors have read and gave final approval for publication.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Fig. S1. Statistics and quality control of full-length transcripts. a Length distribution of subreads. b Length distribution of FLNCs. c length distribution of consensus isoforms using an isoform-level clustering algorithm. d Statistics and classification of full-length transcripts against the G. luofuense reference genome. e Map** read density on the G. luofuense scaffolds; the x-axis represents the scaffold position (Mb), the y-axis represents the median read density (log2), and the green and red lines represent the positive and negative strands of the scaffolds, respectively. f The scale and identity range of all mapped full-length transcripts. The red and blue bars represent the coverage and identity of full-length reads, respectively.
Additional file 2:
Fig. S2. Annotation summary of novel genes from G. luofuense seeds. a The distribution of NR annotations among different seed plant species, the x-axis represents the number of annotated reads. b KEGG enrichment of the annotated novel genes, the x-axis represents the number of annotated reads. c Gene ontology (GO) annotation and categorization of full-length transcripts.
Additional file 3:
Table S1. Detail information in the processing of PacBio sequencing data.
Additional file 4:
Table S2. Detail information in PacBio sequencing data corrected by Illumina sequencing data.
Additional file 5:
Table S3. Summary of annotated numbers of novel genes by the six databases.
Additional file 6:
Table S4. Detail information of Illumina sequenced data from the six G. luofuense seed samples.
Additional file 7:
Table S5. Detail information of genome map** of the transcriptome from the six G. luofuense seed samples.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Deng, N., Hou, C., He, B. et al. A full-length transcriptome and gene expression analysis reveal genes and molecular elements expressed during seed development in Gnetum luofuense. BMC Plant Biol 20, 531 (2020). https://doi.org/10.1186/s12870-020-02729-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-020-02729-1